Difference between revisions of "Popgen"
From CoGepedia
m (→Summary Statistics) |
m (→1. Load a GVCF file) |
||
| Line 9: | Line 9: | ||
== Steps == | == Steps == | ||
===1. Load a GVCF file=== | ===1. Load a GVCF file=== | ||
| − | See the [[LoadExperiment]] tool | + | See the [[LoadExperiment]] tool for loading a GCVF file against an existing genome in CoGe. |
===2. Analyze Diversity=== | ===2. Analyze Diversity=== | ||
Revision as of 11:54, 1 March 2016
CoGe can generate basic population genetics summary statistics given a GVCF input file and an annotated genome.
Contents
Summary Statistics
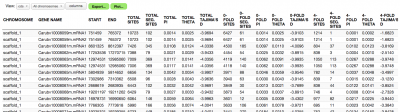
These summary statistics are computed per feature overall (gene, CDS, etc) and by 0/4-fold degeneracy (for CDS only):
- Nucleotide diversity (pi) - average number of pairwise differences between variant sequences
- Watterson's estimator (theta)
- Tajima's D
Steps
1. Load a GVCF file
See the LoadExperiment tool for loading a GCVF file against an existing genome in CoGe.
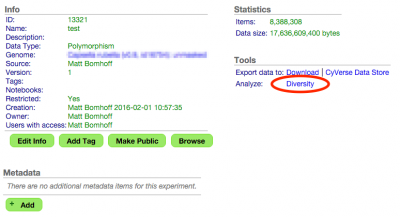
2. Analyze Diversity
In the resulting experiment's menu select "Analyze Diversity".
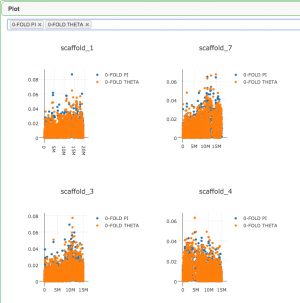
3. Open the finished result
Open the result in the PopGen tool to see results in tabular and graphical form.