Difference between revisions of "Ramosa2 orthologs and CNSs"
| Line 38: | Line 38: | ||
In order to see all connectors between two different genomes hold down shift while clicking on an HSP. If you want to clear the HSP connectors simply hit the “Clear Connectors” button. In addition, when you select an HSP connector a window will pop up showing you more details about the HSP. In order to see the exact alignment simply hit “full annotation” in the pop up window. | In order to see all connectors between two different genomes hold down shift while clicking on an HSP. If you want to clear the HSP connectors simply hit the “Clear Connectors” button. In addition, when you select an HSP connector a window will pop up showing you more details about the HSP. In order to see the exact alignment simply hit “full annotation” in the pop up window. | ||
| + | |||
One of the first things I like to check is that I actually found the genomic region in maize for ramosa2. In order to do this, select an HSP in the mRNA sequence you added by direct submission that corresponds to the maize genomic squence. | One of the first things I like to check is that I actually found the genomic region in maize for ramosa2. In order to do this, select an HSP in the mRNA sequence you added by direct submission that corresponds to the maize genomic squence. | ||
Revision as of 17:16, 15 October 2009
A CoGe Walkthrough – ramosa2 ortholog identification and CNS analysis
In this exercise you will use genomic synteny to identify putative ramosa2 orthologs across several cereal genomes. In addition you will identify Conserved Non-Coding Sequences (CNSs) in the ramosa2 promoter region.
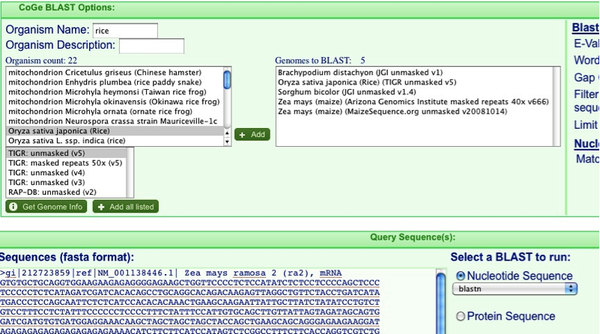
First we need to get some ra2 sequence to use in CoGe Blast. Search the NCBI nucleotide database for: NM_001138446. Copy the Fasta sequence of the mRNA.
Now go to the website for CoGe: http://synteny.cnr.berkeley.edu/CoGe/ (you can just google CoGe and you’ll see it a few pages down). Click on “CoGe Blast” and paste your sequence in the “Sequences field”. It is not necessary to have a Fasta header but it helps keep track of things.
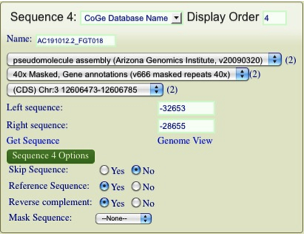
Now we need to decide which organisms we will Blast. Only (mostly) completed genome sequences are available from this window (more on how to add other sequences later on). In the “Organism Name” field search for Sorghum, Rice, Zea mays, and Brachypodium and add them in turn by hitting the “+add” button. I find that it is best to add both the pseudomolecule maize sequence as well as the sequenced BACs as shown in the picture below. Make sure you don’t accidently add the chloroplast genomes instead of the nuclear!
When you’re ready, hit “Run CoGe Blast” to start your analysis.
Note, this is just like Blast, only you are blasting several whole genome sequences at the same time. You can enter protein sequences, change the Blast algorithm and adjust parameters just as you would in any Blast analysis.
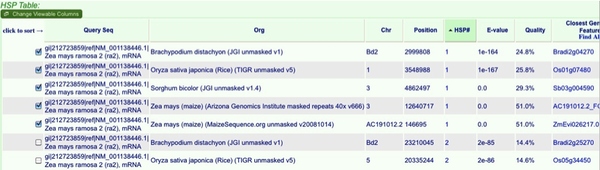
Your results will show both the genomic locations and a table of the “blast hits” or high scoring pairs (HSPs). The HSPs are initially ordered by organism, but you can order them by any of the criteria at the top. To sort HSPs by multiple criteria simply hit one criteria then hold shift while selecting another. I often sort by HSP# then organism. This will give you the top hits in all blasted genomes. You can evaluate the results much like any blast analysis. At this point we will pick which genomic locations to examine for gene synteny. Select the top hit from each genome.
Note that for now we will select top hits from both maize genomes. Even though they are on the same BAC it is often better to see the psuedomolecule assembly as well as the BAC annotations.
Once you have selected the HSPs you would like to analyze, hit “Send to GEvo”. In short, GEvo uses blast to compare the whole genomic regions adjacent to the HSPs you selected.
You’ll notice that you now have a small box for each sequence that you will be comparing. I find that it is helpful to add the original query mRNA sequence to your analysis. To do this hit the “Add Sequence” button at the top left. This will add a blank sequence box. From here you can add any sequence by either direct submission (copy the sequence in!) or by entering the Genbank ID. In this picture I have added the original ra2 mRNA by direct submission.
This function is also helpful when you want to add data from non-sequenced genomes or corrected gene models.
After adding your query sequence, hit “Run GEvo Analysis!” to start.
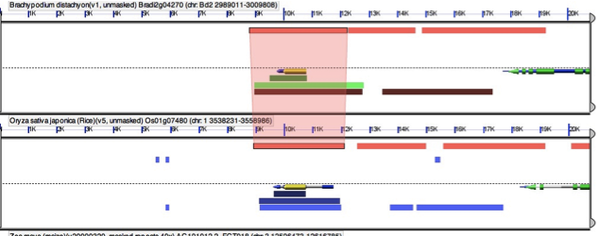
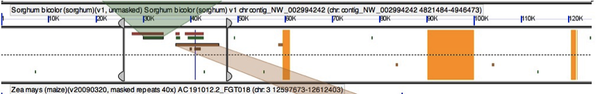
Your GEvo results (http://tinyurl.com/yga5ozu) hit “run GEvo analysis” to recapitulate) will show a small chromosomal region of each genome being compared. Gene models are displayed in the center of each sequence while color coded HSPs are shown on the top and bottom (depending on strand orientation). Gene models are colored in grey (gene), green (CDS), and blue (mRNA). Models that correspond to your query are colored yellow. Each HSP is connected between two genomes. In order to see the connector, simply click on the HSP. For example, this image shows the HSP connector between the coding regions of the putative ramosa2 genes in Rice and Brachypodium.
In order to see all connectors between two different genomes hold down shift while clicking on an HSP. If you want to clear the HSP connectors simply hit the “Clear Connectors” button. In addition, when you select an HSP connector a window will pop up showing you more details about the HSP. In order to see the exact alignment simply hit “full annotation” in the pop up window.
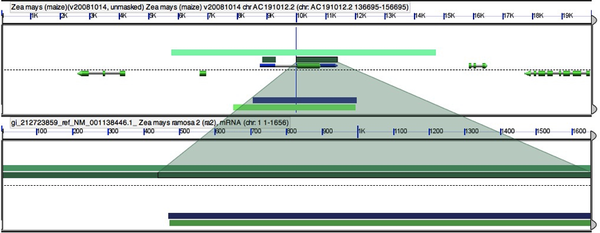
One of the first things I like to check is that I actually found the genomic region in maize for ramosa2. In order to do this, select an HSP in the mRNA sequence you added by direct submission that corresponds to the maize genomic squence.
You can now hit “full annotation” in the HSP pop up window and you will see that the alignment is exact. We found ra2 in the maize genome! Unfortunately it looks like the ra2 gene in the 40x masked psuedomolecule sequence may have been masked out of existence! Maize is a bit of a pain in the shape it’s in. As you poke around you may notice that the orientation of a gene may by wrong. For example in our case Brachypodium and rice are in the opposite orientation to the rest of the sequences.
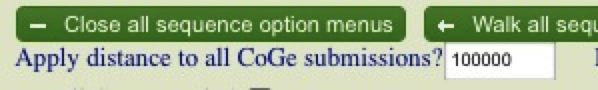
In order to fix this, simply hit the “Reverse Compliment” button in the boxes corresponding to Brachypodium and rice. Hit “Run GEvo Analysis!” to redo the analysis. (http://tinyurl.com/yg9g22q) Now the stage is set to compare synteny between the putative ra2 regions. In order to do this you need to increase the size of the region you are examining. The easiest way to do this is to use the “Apply distance to all CoGe submissions?” By default CoGe compares 10000 bp. Lets increase the regions to 100000. (as an alternative you can increase the region for each individual sequence in the sequence box).