Install coge: Difference between revisions
| Line 167: | Line 167: | ||
# CoGe Configuration File | # CoGe Configuration File | ||
# | # | ||
#database configuration | #database configuration | ||
DB mysql | DB mysql | ||
| Line 175: | Line 175: | ||
DBUSER coge | DBUSER coge | ||
DBPASS 321coge123 | DBPASS 321coge123 | ||
#basic auth name and password | #basic auth name and password | ||
AUTHNAME 6mv9x9lyts8oje8uj3t6yo | AUTHNAME 6mv9x9lyts8oje8uj3t6yo | ||
AUTHPASS f59ba33ee35d363ffefd8b27b375e587b0e5c7a1 | AUTHPASS f59ba33ee35d363ffefd8b27b375e587b0e5c7a1 | ||
# DE public key for JWT in resources/ | # DE public key for JWT in resources/ | ||
DE_PUBLIC_KEY DE_rsa.pub | DE_PUBLIC_KEY DE_rsa.pub | ||
#web cookie name | #web cookie name | ||
COOKIE_NAME cogec | COOKIE_NAME cogec | ||
#support email address | #support email address | ||
SUPPORT_EMAIL coge.genome@gmail.com | SUPPORT_EMAIL coge.genome@gmail.com | ||
#data dir for coge's programs | #data dir for coge's programs | ||
DATADIR /storage/coge/data/ | DATADIR /storage/coge/data/ | ||
#cache dir | #cache dir | ||
CACHEDIR /scratch/coge/cache | CACHEDIR /scratch/coge/cache | ||
#dir for pair-wise whole genome comparisons (e.g. SynMap) | #dir for pair-wise whole genome comparisons (e.g. SynMap) | ||
DIAGSDIR /opt/apache2/coge/web/data/diags/ | DIAGSDIR /opt/apache2/coge/web/data/diags/ | ||
#dir for popgen analysis results | #dir for popgen analysis results | ||
POPGENDIR /storage/coge/data/popgen/ | POPGENDIR /storage/coge/data/popgen/ | ||
#fasta dir | #fasta dir | ||
FASTADIR /opt/apache2/coge/web/data/fasta/ | FASTADIR /opt/apache2/coge/web/data/fasta/ | ||
#sequence dir | #sequence dir | ||
SEQDIR /storage/coge/data/genomic_sequence/ | SEQDIR /storage/coge/data/genomic_sequence/ | ||
#experiment dir | #experiment dir | ||
EXPDIR /storage/coge/data/experiments/ | EXPDIR /storage/coge/data/experiments/ | ||
#temp dir for coge | #temp dir for coge | ||
TEMPDIR /opt/apache2/coge/web/tmp/ | TEMPDIR /opt/apache2/coge/web/tmp/ | ||
#secure temp dir | #secure temp dir | ||
SECTEMPDIR /scratch/coge/tmp/ | SECTEMPDIR /scratch/coge/tmp/ | ||
# IRODS dir | # IRODS dir | ||
IRODSDIR /iplant/home/<USER>/coge_data | IRODSDIR /iplant/home/<USER>/coge_data | ||
IRODSSHARED /iplant/home/shared | IRODSSHARED /iplant/home/shared | ||
IRODSENV /opt/apache/coge/irodsEnv | IRODSENV /opt/apache/coge/irodsEnv | ||
#Base URL for web-site | #Base URL for web-site | ||
URL /coge/ | URL /coge/ | ||
API_URL /api/v1/ | API_URL /api/v1/ | ||
#URL for temp directory | #URL for temp directory | ||
TEMPURL /coge/tmp/ | TEMPURL /coge/tmp/ | ||
#blast style scoring matrix dirs | #blast style scoring matrix dirs | ||
BLASTMATRIX /storage/coge/data/blast/matrix/ | BLASTMATRIX /storage/coge/data/blast/matrix/ | ||
#blastable DB | #blastable DB | ||
BLASTDB /scratch/coge/cache/blast/db/ | BLASTDB /scratch/coge/cache/blast/db/ | ||
#lastable DB | #lastable DB | ||
LASTDB /scratch/coge/cache/last/db/ | LASTDB /scratch/coge/cache/last/db/ | ||
#directory for bed files | #directory for bed files | ||
BEDDIR /opt/apache2/coge/web/data/bed/ | BEDDIR /opt/apache2/coge/web/data/bed/ | ||
#WIKI URL | #WIKI URL | ||
WIKI_URL https://genomevolution.org/wiki/index.php | WIKI_URL https://genomevolution.org/wiki/index.php | ||
#servername for links | #servername for links | ||
#SERVER https://genomevolution.org/coge/ | #SERVER https://genomevolution.org/coge/ | ||
SERVER http://10.140.65.127/coge/ | SERVER http://10.140.65.127/coge/ | ||
#CAS URL | #CAS URL | ||
CAS_URL https://auth.iplantcollaborative.org/cas4 | CAS_URL https://auth.iplantcollaborative.org/cas4 | ||
USER_API_URL https://agave.iplantc.org:443/profiles/v2 | USER_API_URL https://agave.iplantc.org:443/profiles/v2 | ||
MOJOLICIOUS_PORT 3303 | MOJOLICIOUS_PORT 3303 | ||
# Job Engine Server | # Job Engine Server | ||
JOBSERVER localhost | JOBSERVER localhost | ||
# Job Engine Port | # Job Engine Port | ||
JOBPORT 5151 | JOBPORT 5151 | ||
#directory for caching genome browser images | #directory for caching genome browser images | ||
IMAGE_CACHE /opt/apache2/coge/web/data/image_cache/ | IMAGE_CACHE /opt/apache2/coge/web/data/image_cache/ | ||
#maximum number of processor to use for multi-CPU systems | #maximum number of processor to use for multi-CPU systems | ||
MAX_PROC 44 | MAX_PROC 44 | ||
COGE_BLAST_MAX_PROC 8 | COGE_BLAST_MAX_PROC 8 | ||
#True Type Font | #True Type Font | ||
FONT /usr/local/fonts/arial.ttf | FONT /usr/local/fonts/arial.ttf | ||
#various programs | #various programs | ||
BL2SEQ /usr/local/bin/legacy_blast.pl bl2seq | BL2SEQ /usr/local/bin/legacy_blast.pl bl2seq | ||
| Line 295: | Line 295: | ||
CONVERT_BLAST /opt/apache2/coge/bin/convert_long_blast_to_short_blast_names.pl | CONVERT_BLAST /opt/apache2/coge/bin/convert_long_blast_to_short_blast_names.pl | ||
DATASETGROUP2BED /opt/apache2/coge/bin/dataset_group_2_bed.pl | DATASETGROUP2BED /opt/apache2/coge/bin/dataset_group_2_bed.pl | ||
#stuff for Mauve and whole genome alignments | #stuff for Mauve and whole genome alignments | ||
MAUVE /opt/apache2/coge/bin/GenomeAlign/progressiveMauve-muscleMatrix | MAUVE /opt/apache2/coge/bin/GenomeAlign/progressiveMauve-muscleMatrix | ||
COGE_MAUVE /opt/apache2/coge/bin/GenomeAlign/mauve_alignment.pl | COGE_MAUVE /opt/apache2/coge/bin/GenomeAlign/mauve_alignment.pl | ||
MAUVE_MATRIX /opt/apache2/coge/web/data/blast/matrix/nt/Mauve-Matrix-GenomeAlign | MAUVE_MATRIX /opt/apache2/coge/web/data/blast/matrix/nt/Mauve-Matrix-GenomeAlign | ||
# RNA-seq pipelines | # RNA-seq pipelines | ||
PARSE_CUFFLINKS /opt/apache2/coge/scripts/parse_cufflinks.py | PARSE_CUFFLINKS /opt/apache2/coge/scripts/parse_cufflinks.py | ||
# SNP pipelines | # SNP pipelines | ||
PLATYPUS /opt/apache2/coge/bin/Platypus_0.8.1/Platypus.py | PLATYPUS /opt/apache2/coge/bin/Platypus_0.8.1/Platypus.py | ||
GATK /opt/apache2/coge/bin/GenomeAnalysisTK.jar | GATK /opt/apache2/coge/bin/GenomeAnalysisTK.jar | ||
PICARD /opt/apache2/coge/bin/picard-tools-2.4.1/picard.jar | PICARD /opt/apache2/coge/bin/picard-tools-2.4.1/picard.jar | ||
# ChIP-seq pipeline | # ChIP-seq pipeline | ||
HOMER_DIR /opt/apache2/coge/bin/Homer | HOMER_DIR /opt/apache2/coge/bin/Homer | ||
#THIRD PARTY URLS | #THIRD PARTY URLS | ||
GENFAMURL http://dev.gohelle.cirad.fr/genfam/?q=content/upload | GENFAMURL http://dev.gohelle.cirad.fr/genfam/?q=content/upload | ||
Revision as of 17:56, 10 June 2016
Installing CoGe on Ubuntu
Note: these instructions were last updated and verified on June 3rd, 2016.
Initial Dependencies
Run the following command:
sudo apt-get -y install {package}
where {package} is each of the following:
apache2 aragorn blast2 build-essential checkinstall expat gcc-multilib git graphviz imagemagick libdb-dev libgd2-xpm-dev libperl-dev libgd-gd2-perl libconfig-yaml-perl libssl-dev libzmq3-dev mysql-server ncbi-blast+ ncbi-blast+-legacy njplot phpmyadmin python-dev python-numpy python-software-properties samtools swig sqlite3 ttf-mscorefonts-installer ubuntu-dev-tools libapache-asp-perl libapache2-mod-perl2 libapache2-mod-wsgi python-pip nodejs npm libboost-all-dev (for TopHat)
Create MySQL database
Dump CoGe database schema (if using existing CoGe installation, otherwise see schema file below).
mysqldump -d -h localhost -u root -pXXXXXXX coge | sed 's/AUTO_INCREMENT=[0-9]*\b//' > coge_mysql_schema.sql
- CoGe MySQL database schema file (updated June 3rd, 2016): http://genomevolution.org/coge/coge_mysql_schema.sql
- Note: be sure to disable AppArmor for MySQL.
Create new CoGe Database
create database coge
Initialize new coge database
mysql -u root -pXXXXXXXX coge < coge_mysql_schema.sql
Populate a few entries in the feature_type table
- Use the table here which contains 10 feature types: http://genomevolution.org/coge/coge_feature_types.sql
mysql -u root -pXXXXXXXXX coge < coge_feature_types.sql
Create new MySQL user for the CoGe database
use mysql; create user 'coge'@'localhost' IDENTIFIED BY 'XXXXXX'; grant all privileges on coge.* to coge; flush privileges;
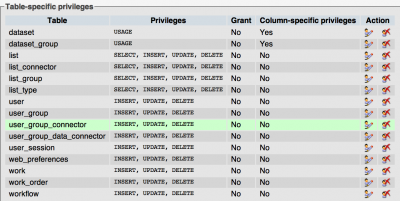
Note: The CoGe web-user needs edit/insert permission on some tables. Here is a snapshot of what these are:
Deploy the Web Site
Generate a public key and add to your GitHub account
See https://help.github.com/articles/generating-an-ssh-key/
Download the CoGe repository
git clone https://github.com/LyonsLab/coge.git
Run setup script to make required subdirectories
cd coge/web ./setup.sh
Configure apache
The /etc/apache2/sites-available/default.conf should look like this:
<VirtualHost *>
ServerAdmin webmasterl@localhost
DocumentRoot /opt/coge/web
<Files *.pl>
SetHandler perl-script
PerlResponseHandler ModPerl::Registry
Options +ExecCGI
PerlSendHeader On
</Files>
<Directory />
Options FollowSymLinks
AllowOverride None
</Directory>
Alias /gobe/ /opt/coge/web/gobe/
<Directory /opt/coge/web/gobe/>
Options +FollowSymLinks +ExecCGI
AddHandler wsgi-script .py
</Directory>
<Directory /opt/coge>
Options Includes ExecCGI FollowSymLinks
AllowOverride All
SetEnv COGE_HOME "/opt/coge/"
Order allow,deny
Allow from all
</Directory>
<Directory /opt/coge/web/services/>
Options +FollowSymLinks +ExecCGI
AddHandler wsgi-script .py
</Directory>
<Directory /opt/coge/web/services/JBrowse/JBrowse_TrackContent_WS/>
Options +FollowSymLinks +ExecCGI
AddHandler wsgi-script .py
</Directory>
ScriptAliasMatch (?i)^/coge/jex(.*) /opt/coge/web/services/jex.py/$1
AliasMatch (?i)^/coge(.*) /opt/coge/web/$1
ProxyPass /coge/api/v1/ http://localhost:3303/
ProxyPassReverse /coge/api/v1/ http://localhost:3303/
ErrorLog /var/log/apache2/error.log
# Possible values include: debug, info, notice, warn, error, crit, alert, emerg.
LogLevel warn
CustomLog /var/log/apache2/access.log combined
ServerSignature On
</VirtualHost>
Enable Required Apache Modules
sudo a2enmod rewrite headers proxy proxy_http expires perl ssl
and reset Apache
Configure coge.conf file
Replacing XXX's with your own information. (Change paths as necessary; this template is configured for having the Coge directory in the path: /opt/coge)
# # CoGe Configuration File # #database configuration DB mysql DBNAME coge DBHOST localhost DBPORT 3307 DBUSER coge DBPASS 321coge123 #basic auth name and password AUTHNAME 6mv9x9lyts8oje8uj3t6yo AUTHPASS f59ba33ee35d363ffefd8b27b375e587b0e5c7a1 # DE public key for JWT in resources/ DE_PUBLIC_KEY DE_rsa.pub #web cookie name COOKIE_NAME cogec #support email address SUPPORT_EMAIL coge.genome@gmail.com #data dir for coge's programs DATADIR /storage/coge/data/ #cache dir CACHEDIR /scratch/coge/cache #dir for pair-wise whole genome comparisons (e.g. SynMap) DIAGSDIR /opt/apache2/coge/web/data/diags/ #dir for popgen analysis results POPGENDIR /storage/coge/data/popgen/ #fasta dir FASTADIR /opt/apache2/coge/web/data/fasta/ #sequence dir SEQDIR /storage/coge/data/genomic_sequence/ #experiment dir EXPDIR /storage/coge/data/experiments/ #temp dir for coge TEMPDIR /opt/apache2/coge/web/tmp/ #secure temp dir SECTEMPDIR /scratch/coge/tmp/ # IRODS dir IRODSDIR /iplant/home/<USER>/coge_data IRODSSHARED /iplant/home/shared IRODSENV /opt/apache/coge/irodsEnv #Base URL for web-site URL /coge/ API_URL /api/v1/ #URL for temp directory TEMPURL /coge/tmp/ #blast style scoring matrix dirs BLASTMATRIX /storage/coge/data/blast/matrix/ #blastable DB BLASTDB /scratch/coge/cache/blast/db/ #lastable DB LASTDB /scratch/coge/cache/last/db/ #directory for bed files BEDDIR /opt/apache2/coge/web/data/bed/ #WIKI URL WIKI_URL https://genomevolution.org/wiki/index.php #servername for links #SERVER https://genomevolution.org/coge/ SERVER http://10.140.65.127/coge/ #CAS URL CAS_URL https://auth.iplantcollaborative.org/cas4 USER_API_URL https://agave.iplantc.org:443/profiles/v2 MOJOLICIOUS_PORT 3303 # Job Engine Server JOBSERVER localhost # Job Engine Port JOBPORT 5151 #directory for caching genome browser images IMAGE_CACHE /opt/apache2/coge/web/data/image_cache/ #maximum number of processor to use for multi-CPU systems MAX_PROC 44 COGE_BLAST_MAX_PROC 8 #True Type Font FONT /usr/local/fonts/arial.ttf #various programs BL2SEQ /usr/local/bin/legacy_blast.pl bl2seq BLAST /usr/local/bin/legacy_blast.pl blastall MULTI_LASTZ /opt/apache2/coge/bin/blastz_wrapper/blastz.py LAST_PATH /opt/apache2/coge/bin/last_wrapper/ MULTI_LAST /opt/apache2/coge/bin/last_wrapper/last.py LAGAN /opt/apache2/coge/bin/lagan-64bit/lagan.pl LAGANDIR /opt/apache2/coge/bin/lagan-64bit/ CHAOS /opt/apache2/coge/bin/lagan-64bit/chaos GENOMETHREADER /opt/apache2/coge/bin/gth DIALIGN /opt/apache2/coge/bin/dialign2_dir/dialign2-2_coge DIALIGN2 /opt/apache2/coge/bin/dialign2_dir/dialign2-2_coge DIALIGN2_DIR /opt/apache2/coge/bin/dialign2_dir/ HISTOGRAM /opt/apache2/coge/bin/histogram.pl KS_HISTOGRAM /opt/apache2/coge/bin/ks_histogram.pl TANDEM_FINDER /opt/apache2/coge/bin/dagchainer/tandems.py DAGCHAINER /opt/apache2/coge/bin/dagchainer_bp/dag_chainer.py EVALUE_ADJUST /opt/apache2/coge/bin/dagchainer_bp/dagtools/evalue_adjust.py FIND_NEARBY /opt/apache2/coge/bin/dagchainer_bp/dagtools/find_nearby.py QUOTA_ALIGN /opt/apache2/coge/bin/quota-alignment/quota_align.py CLUSTER_UTILS /opt/apache2/coge/bin/quota-alignment/cluster_utils.py BLAST2RAW /opt/apache2/coge/bin/quota-alignment/scripts/blast_to_raw.py SYNTENY_SCORE /opt/apache2/coge/bin/quota-alignment/scripts/synteny_score.py CODEML /opt/apache2/coge/bin/codeml/codeml-coge CODEMLCTL /opt/apache2/coge/bin/codeml/codeml.ctl CONVERT_BLAST /opt/apache2/coge/bin/convert_long_blast_to_short_blast_names.pl DATASETGROUP2BED /opt/apache2/coge/bin/dataset_group_2_bed.pl #stuff for Mauve and whole genome alignments MAUVE /opt/apache2/coge/bin/GenomeAlign/progressiveMauve-muscleMatrix COGE_MAUVE /opt/apache2/coge/bin/GenomeAlign/mauve_alignment.pl MAUVE_MATRIX /opt/apache2/coge/web/data/blast/matrix/nt/Mauve-Matrix-GenomeAlign # RNA-seq pipelines PARSE_CUFFLINKS /opt/apache2/coge/scripts/parse_cufflinks.py # SNP pipelines PLATYPUS /opt/apache2/coge/bin/Platypus_0.8.1/Platypus.py GATK /opt/apache2/coge/bin/GenomeAnalysisTK.jar PICARD /opt/apache2/coge/bin/picard-tools-2.4.1/picard.jar # ChIP-seq pipeline HOMER_DIR /opt/apache2/coge/bin/Homer #THIRD PARTY URLS GENFAMURL http://dev.gohelle.cirad.fr/genfam/?q=content/upload
Install Perl Modules
- Install cpanminus
sudo cpan install App::cpanminus
- Install third-party modules required by CoGe
cat modules.txt | xargs sudo cpanm
- Install CoGe-specific modules
./make_perl.sh
- After installing modules, reset the Apache webserver
sudo service apache2 restart
Install Python Modules
sudo pip install pyzmq
Install Javascript dependencies
- Install javascript dependencies
sudo ln -s /usr/bin/nodejs /usr/bin/node sudo npm install -g bower bower install
Install Third-Party Bioinformatics Tools
Download the programs listed below and follow the installation instructions on their respective websites.
Most programs can be installed with the following commands (but check the documentation for each program):
./configure --prefix=/usr/local/ make sudo make install
- GSNAP/GMAP: http://research-pub.gene.com/gmap/
- FastBit: https://sdm.lbl.gov/fastbit/
- Clustalw: http://www.clustal.org/clustal2/
- Bowtie: http://bowtie-bio.sourceforge.net/bowtie2/index.shtml
- TopHat: http://ccb.jhu.edu/software/tophat/index.shtml
- HISAT2: https://ccb.jhu.edu/software/hisat2/index.shtml
- Cufflinks: http://cole-trapnell-lab.github.io/cufflinks/
- Nwalign: https://pypi.python.org/pypi/nwalign/?
- Cutadapt: http://cutadapt.readthedocs.io/en/stable/installation.html ... sudo su ; sudo pip install cutadapt
- TrimGalore: http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/
- Picard: http://broadinstitute.github.io/picard/ ... requires Java 8
- Platypus: http://www.well.ox.ac.uk/platypus
- HSTlib (required by Platypus): http://www.htslib.org/download/
- Lastz: download the tarball http://www.bx.psu.edu/~rsharris/lastz/ then edit the src/Makefile and remove the word -Werror from line 31. Then run make and make install.
- Last aligner (v731 or greater is required): http://last.cbrc.jp/
- VCFTools: https://github.com/vcftools/vcftools
- Vcfutils: https://github.com/lh3/samtools/blob/master/bcftools/vcfutils.pl
- EMBOSS (sizeseq program): http://emboss.sourceforge.net/ ... Run "sudo ldconfig" after "make install"
- iCommands: https://github.com/irods/irods-legacy (OPTIONAL: only required if CyVerse authentication and Data Store services are available; note that the legacy version is required, not the latest)
- Bismark: http://www.bioinformatics.babraham.ac.uk/projects/bismark/
- BWAmeth: https://github.com/brentp/bwa-meth
- Homer: http://homer.salk.edu/homer/ ... sudo perl ./configureHomer.pl -install homer
- Blat (required by Homer): https://genome.ucsc.edu/FAQ/FAQblat.html
Install Third-Party Fonts
Download from here: https://www.microsoft.com/typography/fonts/font.aspx?FMID=1705
And copy to /usr/local/fonts/arial.ttf (or whatever path you set in the coge.conf config file under FONT)
Install blast matrices
cd /storage/coge/data/blast git clone https://github.com/LyonsLab/blast-matrix.git mv blast-matrix matrix
Install JBrowse
Copy from existing CoGe installation if one exists. Otherwise, download and install the JBrowse package from http://jbrowse.org/install/
unzip JBrowse-1.11.4-dev.zip mv JBrowse-1.11.4 /coge/web/js/jbrowse
Install CCTools
- Download a stable release that is 4.3 or greater from http://ccl.cse.nd.edu/software/downloadfiles.php
- Extract the file (this example is using version 4.3 which may differ from the version downloaded)
tar xzvf cctools-4.3.0-source.tar.gz
- Compile and install
cd cctools-4.3.0-source ./configure --prefix /usr/local make sudo make install
- Add the following upstart scripts for the work_queue_pool and catalog_server to /etc/init
By default the pool directory for work_queue will be in /storage/work_queue adjust the directory as needed.
# /etc/init/.conf
description "The cctools work queue pool"
author "Evan Briones"
start on (local-filesystems and net-device-up IFACE=eth0)
stop on shutdown
respawn limit 30 60
pre-start script
POOL_DIR=/storage/work_queue
LOG_FILE=$POOL_DIR/logs/work_queue_pool.log
# Add the pool directory and set ownership
if ! [ -d "$WORK_DIR" ]; then
mkdir -p $POOL_DIR/workers
mkdir -p $POOL_DIR/logs
chown -R www-data:www-data $POOL_DIR
fi
# Remove the pidfile if it exists
rm -f $POOL_DIR/work_queue_pool.pid
# Archive old log and timestamp the value
if [ -f "$LOG_FILE" ]; then
TIMESTAMP=$(date +"%Y-%m-%d.%H.%m.%S")
mv -f $LOG_FILE "$LOGFILE.$TIMESTAMP"
fi
end script
script
POOL_DIR=/storage/work_queue
LOG_FILE=$POOL_DIR/logs/work_queue_pool.log
#CONFIG=/etc/yerba/work_queue_pool.conf
#WORK_QUEUE_POOL=$(which work_queue_pool)
WORK_QUEUE_FACTORY=$(which work_queue_factory)
export CATALOG_HOST=localhost
export CATALOG_PORT=1024
exec start-stop-daemon -c www-data -g www-data -d $POOL_DIR --start \
-p $POOL_DIR/work_queue_pool.pid --exec $WORK_QUEUE_FACTORY \
-- -T local -M coge-main -d all -o $LOG_FILE -w 10 \
-S $POOL_DIR -E "--workdir=$POOL_DIR/workers"
end script
# /etc/init/.conf
description "The cctools catalog server"
author "Evan Briones"
start on (local-filesystems and net-device-up IFACE=eth0)
stop on shutdown
respawn limit 30 60
script
exec catalog_server -p 1024 -l 100 -T 3
end script
- Start the catalog server and work_queue_pool
sudo start work_queue_pool sudo start catalog_server
Install the Job Engine (Yerba)
Download and install the latest Yerba package from https://github.com/LyonsLab/Yerba/archive/v0.3.4.tar.gz
For more specific details on Yerba visit https://github.com/LyonsLab/Yerba/
The default installation path for Yerba will be in /opt/Yerba. If another path is chosen update the configuration files to match.
- Copy and the configuration file to /etc/yerba/yerba.cfg
[DEFAULT] debug = True access-log = /opt/Yerba/log/access.log yerba-log = /opt/Yerba/log/yerbad.log [yerba-log] logging = /etc/yerba/logging.conf [access-log] logging = /etc/yerba/access.conf [yerba] port = 5151 level = DEBUG [workqueue] catalog_server = localhost catalog_port = 1024 project = coge-main log = /var/log/workqueue.log port = -1 password = /etc/yerba/workqueue_pass debug = True [db] path = /opt/Yerba/workflows.db start_index = 100
- Copy the upstart file to /etc/upstart/yerba.conf
# /etc/init/yerba.conf
description "Yerba server daemon"
author "Evan Briones"
start on (local-filesystems and net-device-up IFACE=eth0)
stop on shutdown
respawn
pre-start script
LOG_DIR=/opt/Yerba/log
LOG_FILE=$LOG_DIR/debug.log
[ -d "$LOG_DIR" ] || mkdir -m777 -p $LOG_DIR
# [ -f "$LOG_FILE" ] || rm -f $LOG_FILE
end script
script
export YERBA_ROOT=/opt/Yerba
export PYTHONPATH="/usr/local/lib/python2.7/site-packages:$YERBA_ROOT"
exec start-stop-daemon -c www-data -g www-data --start \
--iosched real-time --nicelevel -19 \
--exec $YERBA_ROOT/bin/yerbad -- >> $YERBA_ROOT/log/debug.log 2>&1
end script
post-start script
echo Restart on: `hostname -A` | mail -s "UPSTART: Yerba was started" coge.genome@gmail.com
end script
- Initialize and start the job engine
/opt/Yerba/bin/yerbad --setup sudo chown www-data:www-data /opt/Yerba/workflows.db sudo start yerba
Troubleshooting
Visualization in GEvo does not work
This relies on a system known as Gobe. Check the following things:
- Apache configuration for gobe
- Make sure the Python Web module is installed: sudo aptitude install python-webpy
- Check to see if paths hard-coded into gobe/flash/service.wsgi need to be updated
- NOTE: Not sure if this is required
Working on an Atmosphere Virtual Machine
Click here for instructions on dealing with issues that occur specifically with Atmosphere Virtual machines.