Difference between revisions of "Insertion"
From CoGepedia
| Line 1: | Line 1: | ||
Insertion can be thought of as opposite of [[deletion]] mediated by transposase. A transposase will cut the genome at a specific cleveage site creating staggered nicks followed by insertion of transposon or a linear piece of DNA. The gaps in the genome are then reparied and ligated which results in direct repeats bordering the transposon insertion. Note that the insertion in a genome becomes evident by the presence of direct repeats at the edges of inserted genes which are not usually expected to be found in a particular genome or at a particular locus in a genome. | Insertion can be thought of as opposite of [[deletion]] mediated by transposase. A transposase will cut the genome at a specific cleveage site creating staggered nicks followed by insertion of transposon or a linear piece of DNA. The gaps in the genome are then reparied and ligated which results in direct repeats bordering the transposon insertion. Note that the insertion in a genome becomes evident by the presence of direct repeats at the edges of inserted genes which are not usually expected to be found in a particular genome or at a particular locus in a genome. | ||
| − | [[Image:insertion.png|500px|thumb|center| Image taken from: [http://www.biology.ucsd.edu/classes/old.web.classes/bimm100.FA00/09. | + | [[Image:insertion.png|500px|thumb|center| Image taken from: [http://www.biology.ucsd.edu/classes/old.web.classes/bimm100.FA00/09.MobileElements.html#A] ]] |
Latest revision as of 17:55, 12 November 2009
Insertion can be thought of as opposite of deletion mediated by transposase. A transposase will cut the genome at a specific cleveage site creating staggered nicks followed by insertion of transposon or a linear piece of DNA. The gaps in the genome are then reparied and ligated which results in direct repeats bordering the transposon insertion. Note that the insertion in a genome becomes evident by the presence of direct repeats at the edges of inserted genes which are not usually expected to be found in a particular genome or at a particular locus in a genome.
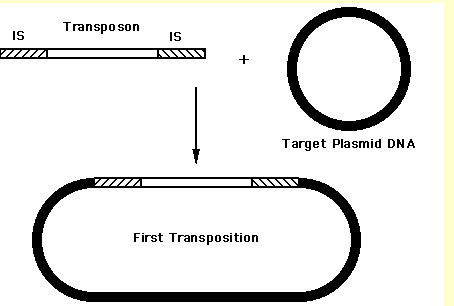
Image taken from: [1]