UGTs through the genus Brassica
Contents
UGT Project
Introduction
The genus Brassica
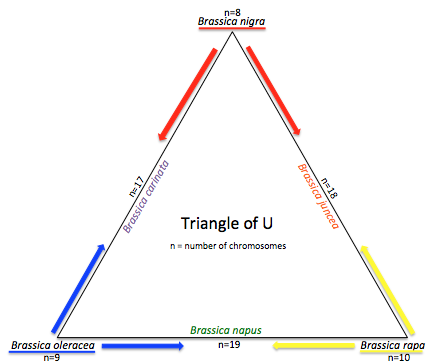
The genus Brassica consists of over thirty wild species and hybrids, or morphotypes. Generally, species from the genus Brassica are crop species like broccoli, cauliflower, cabbage, mustard and more. The Brassica genome has undergone more polyploidy events than A. thaliana. A. thaliana is notable for being a model organism because of its extensive genetic maps of the 5 chromosomes, prolific seed production aired with the fact that it has a relatively small genome (TAIR, About Arabidopsis). The genus Brassica has undergone two tetraploidy and two hexaploidy events, one more than Arabidopsis, since the eudicot paleohexaploidy event, which gave rise to Vitis, Prunus, Arabidopsis, and Brassica (Figure 1). Brassica saw one more triplication event after Arabidopsis. Had there been no gene loss, the ratio of genes in Arabidopsis thaliana to Brassica rapa would be 1:3. The “Triangle of U” theory describes the genetic relationship between six species of Brassica: Brassica rapa, Brassica nigra, Brassica oleracea, Brassica juncea, Brassica carinata, and Brassica napus. B. juncea, B. carinata, and B. napus are all allotetraploids, hybrids with four times the chromosome set of haploids (Figure 2).
UGT functions
Uridine diphosphate (UDP) glycosyltransferases (UGTs) mediate transfer of glycosyl residues from activated nucleotide sugars to acceptor molecules (Tang, Unleashing the Genome of the Brassica rapa). They provide instructions for making enzymes that perform the process of glycosylation, the addition of glucuronic acid to a substrate (Genetics Home Reference, UGT gene family). This pathway is particularly important in metabolism, and many regard the UGT enzyme as the most important enzyme in the pathway. In humans, these enzymes are responsible for the breakdown of several prescription drugs (Glycosyltransferases) Plant UGTs are also known to contribute cellular homeostasis and the detoxification of xenobiotics by metabolizing the pollutants from pesticides.
UGT chemistry
By mediating transfer of glycosyl residues from activated nucleotide sugars to acceptor molecules, UGTs regulate properties of those acceptors such as bioactivity, solubility and transport within cells and throughout organisms (Ross, Higher plant glycosyltransferases). The UGT enzymes are involved in the metabolic process of glycosylation during phase II metabolism. Drug metabolism is separated into three phases, the second of which involves the UGT enzymes.
Purpose
UGTs are vital to metabolism of all organisms. Several UGT genes of Arabidopsis thaliana have been sequenced already. Looking into the sequences of Arabidopsis lyrata and Brassica rapa, I hope to determine the exact ratio of UGT genes in each of the species, discover similarities among phylogenetic trees, and pinpoint which genes were conserved, lost, and altered.
Preliminary Data
Of the 28 Glycosyltransferase Families that The Arabidopsis Information Resource (TAIR https://www.arabidopsis.org/) has on A. thaliana, I chose to work with Family 1 due to the fact that most of the genes were similar in function as the TIGR Annotation suggested. The FASTA sequence for gene AT5G65550 was used as a query sequence in the JGI Phytozome database to recover A. lyrata genes (Phytozome https://phytozome.jgi.doe.gov/pz/portal.html). The Brassica Database (BRAD https://brassicadb.org/) was used to recover orthologs for the B. rapa genes. A table was developed to organize the information collected from each database (Table 1). A preliminary tree was constructed of 114 of these genes which CoGe had FASTA sequences for (Figure 3 [to be created: PDF]). Several functions of the genes in the preliminary tree were identified and noted (Functional Genomics). Three clusters on the tree were chosen by random sampling and named Test groups. Test group 1 genes exhibit functions of flavonol 7-O-glucosyltransferase and brassinosteroid O-glucosyltransferase. Test group 2 genes exhibit the function of having ABA glucosyltransferase activity. Test group 3 genes exhibit the function of monolignol 4-O-glycosyltransferase activity and having xeniobiotic glycosyltransferase activity. Each of these test groups genes’ FASTAs were placed into one file. An inventory of the test groups was organized (Table 2). Using GEvo, each A. thaliana gene was visualized for syntenic regions in A. lyrata and B. rapa. The FASTAs for those genes in were added to the file and the GEvolinks were saved to a separate files for each test group. Using phylogeny.fr, new trees were constructed with A. thaliana, A. lyrata, and B. rapa for each test group (Figures 4, 5, 6).
Hypotheses
UGTs are found in all living organisms. The unique opportunity presented by the species A. thaliana, A. lyrata, and B. rapa allows for plant UGTs to be characterized and tracked through the highly duplicated Brassicas. Doing so may uncover more specific functions on certain UGT genes and track how functions are conserved or altered over evolutionary history. With these possibilities in mind, hypotheses were developed to delve further into the genetic information provided by the species.
H0: The evolutionary history of Brassica rapa reveals no new information regarding UGT function.
H1: Brassica rapa UGT genes are redundant and found three times as much as the UGTs found in Arabidopsis thaliana and Arabidopsis lyrata.
H2: Brassica rapa UGT genes have more inversions and polymorphisms than those in Arabidopsis lyrata compared to Arabidopsis thaliana.
Methods
Python Code
A simple program called FindDifferences.py was developed to isolate the A. thaliana genes that are found in both TAIR’s Glycosyltransferase Family 1 list and CoGe and those found just in the same TAIR list. Any genes found only in CoGe were removed from the list of A. thaliana UGT genes. The program takes in two csv files: one containing the genes from TAIR and one containing the genes from. The program reads each file, changes each gene name to one format using strip functions, then compares each file and returns which gene names are only in one list and prints the list to a new csv. [Insert GitHub link to program].
CoGe BLAST
The BLAST tool in CoGe uses a query sequence of amino acids to find other genes or contigs with syntenic regions in A. thaliana. The query sequence, AT5G65550, for the BLAST search used to construct the preliminary tree was chosen randomly from the Glycosyltransferase Family 1 genes of TAIR. This process was repeated several times. The same query sequence was used in BLAST for A. lyrata genes and B. rapa genes. A second query sequence of amino acids, Bra037821, was used from a B. rapa gene orthologous to the A. thaliana gene.
FASTA View
The FASTA View feature in CoGe provides FASTA files for each gene inputted. From CoGe BLAST, selected genes and can be sent to FASTA View where DNA and protein sequences can be retrieved. 122 protein sequences of the 126 found in CoGe BLAST were retrieved for the preliminary tree of A. thaliana. This pool of genes was used for the tree construction.
Phylogeny.fr
Phylogeny.fr is a tree-rendering program for which inputs in the format of FASTA files are used to output a phylogenetic tree. This program has been used to construct four trees so far. The preliminary tree of Arabidopsis thaliana genes consisted of 114 genes showing some interesting tandem duplications.
Test Groups
From the preliminary tree, three test groups were isolated by random sampling (see Table 2).
SynFind
For each gene within each test group, the SynFind tool in CoGe was used to find syntenic regions among all three species, A. thaliana, A. lyrata, and B. rapa. The tool takes uses a gene from A. thaliana and finds regions of synteny in the target genomes selected. SynFind returns a table of gene matches with synteny scores. From this result, several tools and features can be accessed, like GEvo, which was used in this instance.
GEvo
Two steps were taken to continue the research after finding the gene matches. The FASTA sequences were generated using FASTA View and saved to one file. In addition, the gene matches were compared and visualized in GEvo. The GEvo tool aligns the sequences of each gene match and shows regions of synteny with color coordinated boxes. Visualizing in GEvo allows for true matches to be found and others which are just noise to be removed. For the construction of the three new trees, no noise was removed. Links to each visualization were saved to a file.