Using CoGe for the analysis of Plasmodium spp
1. Finding and inputing data into CoGe
1.1 Finding about the Plasmodium spp. genomes present in CoGe
The number of Plasmodium genomes available to the public increases yearly. Numerous research groups are working on completing the Plasmodium genome panorama, leading to reposition of diverse genome sequences under diverse levels of completion and originating from a variety of databases. A large number of Plasmodium genomes have been deposited on the National Center for Biotechnology Information (NCBI); however, additional databases such as PlasmoDB ([1]), GeneDB ([2]) and MalAvi ([3]) also carry addiional Plasmodium genome sequences.
In order to attain a better picture of Plasmodium spp. genome evolution, the CoGe platform can be used to perform diverse comparative analyses. Currently, there is a number of Plasmodium genomes available on the CoGe database. You can obtain more about them by following these steps:
1. Go to: [[4]]
2. Create an account/ login into CoGe
3. On the main CoGe page, find the Tools tile and click on to Organism View ([5])
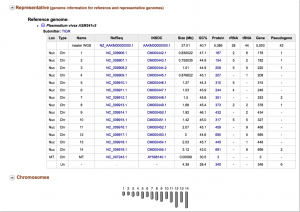
4. Organism View allows the researcher to find all publicly available genomes uploaded into CoGe and browse any corresponding information. You can find any published genome by typing a scientific name into the Search box. For each organism uploaded to CoGe you will find the following information:
- Organisms: In the case of Plasmodium spp., the different parasitic strains currently uploaded. Any organelle genomes independently uploaded (mitochondrial and apicoplast) can also be found here.
- Organism Information: provides an outline of organisms’ taxonomy (following that published on NCBI), quick links to some of the main CoGe analysis tools, and the search engines were information can be found.
- Genomes: All the genome versions for this species. Selecting different genome versions modifies al other output observed in this page; in addition, it allows the user to access to previous versions of a published genome (e.g. access scaffolds from a previous genome version currently under the chromosome assemble level).
- Genome information: Shows the genome IDs, type of sequences uploaded and length of the whole genome. In addition, this tab allows the user to directly perform analyses using the CoGe platform.
- Datasets: This section will show the number of datasets included for this genome. In the case of completely sequenced Plasmodium genomes, this will indicate the code numbers for the datasets of each individual chromosome.
- Dataset information: Provides specific information for each individually selected dataset including. Information includes the accession numbers (if available), source of the upload, chromosome length and GC%.
- Chromosomes: Shows the number of available chromosome for the selected genome. However, depending of the methodology used to upload the data into CoGe and the nature of the dataset itself, the count and length of chromosomes shown will be larger than expected (e.g. will show the number of contigs in lieu of the number of chromosomes). For whole sequenced genomes, specific IDs under the Dataset section will showcase the chromosome number and length.
- Chromosome information: Shows the chromosome ID and the number of base pairs for that chromosome.
5. Under Genome Information, clicking on the Genome Info section permits the user to access to a more detailed genome description. It also allows access to other quick links to comparative analysis tools available on CoGe.
1.2 Uploading Plasmodium spp. genomes into CoGe
While data can be uploaded into CoGe using a variety of methods, we will focus on the two most relevant for the incorporation of Plasmodium spp. genomes. We will follow each method with an example. For additional information, please check the following link: [[6]]
- Uploading genomes from NCBI/Genebank:
- Depending on the researcher's interests, it might be desired to perform analyses using complete Plasmodium genomes or focus only in specific organelles and chromosomes. In order to upload a complete Plasmodium genome, make sure to follow these steps:

- 1. In the upper part of the screen, find the Representative Genome section. Below, the Download Sequences in FASTA format and Download Genome Annotation sections can be found.
- - To download the complete Plasmodium vivax genome, click on Genome under Download Sequences in FASTA
- - To download the complete annotation for the Plasmodium vivax genome, clich on GFF under Download Genome Annotation
- 2. Both files will be downloaded to your desired folder into your local computer.

- 3. Go to: [[7]]
- 4. Login into CoGe.
- 5. Click on the MyData section on the upper left part of the screen.
- 6. This will lead to the Data section of your personal CoGe page. This section will fill up as genomes of interest are uploaded into CoGe.
- 7. On the upper left section of the screen, click the NEW button and select New Genome from the dropdown menu.

- 8. Once on the page to Create a New Genome into CoGe, information about the organisms taxonomy and the genome's origin must be inputed. Depending of the type of organism being uploaded, taxonomic information might have not been yet included into CoGe. If this is the case, a new organisms should be created. To do this the following steps should be followed:
- a. Click on NEW on the "Organism:" section
- b. On the Search NCBI box type the scientific name of the organism to be uploaded. If the organism of interest is not on NCBI, select the closet taxonomic relative. In the case of Plasmodium several strains might be available for a single species, make sure to select the correct strain or, if a new strain is being uploaded, to add the new strain name.
- c. Click Create
- 9. Once the new strain/genome has been added, additional information should be included as well. Depending on the number of genome versions for the selected genome available at CoGe, a different number will be typed on Version. Thus, it is important to check the number of genome version already available on CoGe before inputing a new version. Under the section named Type, select the adequate sequence type from the drop menu (most sequences can be identified as unmasked, [[8]]). Select the Source from the next dropdown menu (in this case NCBI, but there are many other sources available including Private sources). Check if you desire your genome to be Restricted or not.
- - Restricted genomes can only be seen and analyzed by the user and those to whom it has been shared.
- - Unrestricted genomes are available for the general public
- 10. Once done click Next
- 11. Genome files themselves can be uploaded in this window using four different strategies: first, data can be uploaded directly from the Cyverse Data Store (if the data is not on the Data Store, it can be easily uploaded there afterwards once it has been included in CoGe); second, creating an HTP/FTTP link directly to the data; third, Upload the data from a private computer, and fourth, uploading the data using NCBI/GenBank accession numbers. In the following example, the data will be uploaded using the Upload option.
- 12. Select the downloaded file
File:Uploadingusingdownload.png
- 13. bveorbo
- 14. bveorbo
- 15. bveorbo
- 16. bveorbo
- 17. bveorbo
- This same procedure can be used to upload genomes in later stages of assembly (e.g. Scaffolds) into CoGe.
- Now, it is also possible to specifically upload chromosomes and organelles's genomes into CoGe. The following steps show how to upload individual chromosomes into CoGe:
- 1. bgfjyt
- 2. grehe