Finding and intregating Plasmodium genomes to CoGe
Finding and integrating Plasmodium genomes in CoGe
An increasing number of Plasmodium genomes have been sequenced in recent years, a number that will likely increase in the future. Thus, tools that permit rapid integration of genomic information and its subsequent analysis are essential for Plasmodium research. Online platforms aid in reducing computational time, costs, and foment worldwide collaborations. CoGe is one of these platforms.
The first step in analyzing Plasmodium genomes with CoGe is determining which genomes are already included in the data repository.
RETURN TO THE MAIN PAGE: Using_CoGe_for_the_analysis_of_Plasmodium_spp
Finding the Plasmodium genomes already present in CoGe

A significant accomplishment in the study of Plasmodium genomics was the full sequencing and assembly of the P. falciparum genome [1]. Over the years, this genome has been revised and re-annotated, resulting in different "releases", or versions of the P. falciparum genome. CoGe’s repositories contain each of these releases with a unique version identifier (i.e., v5, v4, etc). This happens because the CoGe platform incorporates new versions of a genome without deleting previous ones. Thus, you can find the initial P. falciparum sequenced genome loaded onto CoGe (v3) alongside the more current releases (v5).
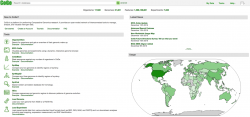
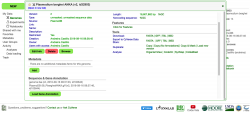
Before importing a genome into CoGe, and to prevent redundancy of genomic information, it is recommended to identify what data has previously been imported. You can search CoGe’s Plasmodium genomes by typing the word "Plasmodium" into the Search bar at the top of most pages (Figure 1). This will retrieve all organisms and genomes with names matching the search term. For instance, when searching the term "plasmodium falciparum 3D7", you will see that there are currently eight publicly available genomes associated with this specific strain of P. falciparum. Clicking on any organism will produce the details of the upload. Alternatively, you can find the Tools section on the main CoGe page (Figure 2) and click on OrganismView (https://genomevolution.org/coge/OrganismView.pl).
All publicly available genomes imported into CoGe, and their corresponding metadata, can be found in OrganismView. To search for any genome on OrganismView, type a scientific name into the Search box. The following information will be displayed (Figure 3):
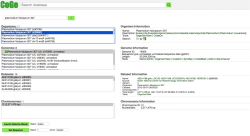
- Organisms: In the case of Plasmodium spp., the different parasitic strains are already imported. In addition, organellar genomes (mitochondrial and apicoplast) have also been imported.
- Organism Information: An outline of the organism's taxonomy (as published on NCBI/Genbank). This section also includes links to some of CoGe's main analysis tools.
- Genomes: All genome versions available. Note that by selecting different genome versions, all associated genomic information changes.
- Genome information: Includes genome IDs, type of sequences uploaded, and sequence length. You can also access CoGe's genome analysis tools in this section.
- Datasets: This section includes the number of datasets for the specified genome. In the case of completely sequenced genomes imported from NCBI/GenBank, it will indicate the chromosome’s accession numbers.
- Dataset information: Provides information for each dataset including accession numbers (if available), the source of the import, chromosome length, and GC%.
- Chromosomes: Shows the number of chromosome in the selected genome. However, depending on the method used to import the genome into CoGe and the dataset itself, the number and length of the chromosomes will vary.
- Chromosome information: Shows each chromosome's ID and lenght on base pairs (bp).
You can find a more detailed description of any genome by accessing the Genome Info section within Genome Information. You can also access links to the majority of CoGe’s comparative analysis tools in this section. Keep in mind that genomes imported to CoGe can be made “Public” or “Restricted”. Genomes made “Public” can be seen and analyzed by anyone using the CoGe platform. “Restricted” genomes can only be seen and/or analyzed by the user and shared accounts (Sharing_data).
Importing Plasmodium genomes into CoGe
If a genome is not found on CoGe's repository then it must be imported before analysis. Genomic data can be imported into CoGe using a variety of methods. We will focus on the two methods most likely to be used when importing genomes. For additional information about other methods please see How_to_load_genomes_into_CoGe. Depending on your intended analyses, you might want to use a complete Plasmodium genome, a specific chromosome, or focus on an organelle. The methods described here can be used to upload either of these data. To import a genome onto CoGe follow these steps:
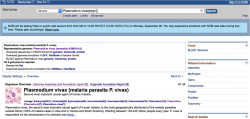
- 1. Go to the genome database on NCBI/GenBank (or your favorite database) and type "Plasmodium" in the search box.
- 2. In the Representative Genome section you will find links to Download Sequences in FASTA format and Download Genome Annotation (Figure 4).
- - To download a complete Plasmodium genome click on Genome under Download Sequences in FASTA.
- - To download a complete annotation for a Plasmodium genome click on GFF under Download Genome Annotation.
- You can also download single chromosomes and, if available, organellar genomes by clicking on their respective RefSeq or INSDC numbers.
- 3. Go to CoGe and log in. You can follow this link: https://genomevolution.org/coge/
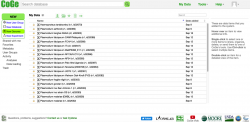
- 4. Click on MyData to reach the Data section of your personal CoGe page (Figure 5). This section will fill up as you import genomes and load Experiments into CoGe.
- 5. Click on NEW and select New Genome from the dropdown menu.
- 6. Input information about the organism's taxonomy and the genome's source on the Create a New Genome window (Figure 6). Consider that taxonomic information for that genome might not have been incorporated into CoGe yet. If this is the case, follow these steps to create a "new organism":
- a. Click on NEW on the "Organism:" section.
- b. Type the scientific name of the organism to be imported on the Search NCBI box. If the organism does not show up select its closest taxonomic relative. In the case of Plasmodium, several strains might be available for a given species (particularly P. vivax and P. falciparum). Make sure to select the correct strain or, if a new strain is being imported, to add its name.
- c. Click Create.
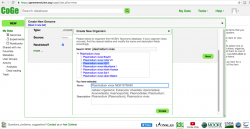
- 7. After creating a new strain/genome, you must also include the import’s metadata. Type the import's genome version in Version after confirming which genome versions are available on CoGe. If this if the first genome imported, the version number should be “1”. Select the sequence type from the dropdown menu on the Type section. Most sequences can be identified as unmasked (check this wiki’s Masked section for further details). Select the Source in the next dropdown menu (in this case NCBI). Finally, tick the check box if you desire your genome to be Restricted.
- 8. Click Next.
- 9. Genome files can be imported to CoGe using four different strategies: 1) import directly from the CyVerse Data Store; 2) create a direct HTP/FTTP link to the data; 3) import the files from a private computer using Upload; and 4) use GenBank accession numbers.
- To import genomes using Upload:
- a. Select a genome file from your local computer and wait for it to be read by CoGe. Once the process is completed select Next.
- b. Click Start to begin the import.
- c. When the import has concluded, the file’s metadata will be visible in the Genome Information page.
- d. To import annotation data click on Load Sequence Annotation under the Sequence & Gene Annotation menu. Note that any upload can be updated at any point. Thus, genome annotations or experimental data can be added later to any genome already in CoGe.
- e. In the Describe your annotation page, select the version and source of the annotation data and click Next. The data can be uploaded from the CyVerse Data Store, by creating a HTTP/FTP link, or by using the Upload option. Once concluded, the genome annotation should be visible on the Genome Information page under the Sequence & Gene Annotation menu (Figure 7). For more details about uploading genome annotations please check LoadAnnotation.
- To import genomes using NCBI/Genebank:
- a. Select the GenBank accession numbers option. Type or Copy/Paste the RefSeq or INSDC numbers for each chromosome or organelle and click Get. Information from each imported genome should appear under Selected file(s). Once all genomes have been imported (e.g. the 14 Plasmodium chromosomes) click on Next.
- b. Once the import has concluded, the file’s metadata will be visible in the Genome Information page. Note that NCBI/GenBank genome annotations will be automatically imported to CoGe when using this method and that genomes uploaded using this method will be automatically made “Public”.
Exporting genomes from CoGe to CyVerse
- Data can be exported into CyVerse for easy sharing and storage after it has been imported onto CoGe. While this is not required to use any of CoGe's tools, it is a recommended step. You can export data from CoGe into the CyVerse Data Store by following these steps:
- 1. While logged into CoGe, go to the genome's Genome Information page.
- 2. Under the Tools menu, find the Export to CyVerse Data Store option. Click either on the FASTA or the GFF file options to upload genomic data and/or its annotation.
- 3. Wait until the export is completed. From this point forward, your FASTA and GFF files will also be found in the CyVerse Data Store.
RETURN TO THE MAIN PAGE: Using_CoGe_for_the_analysis_of_Plasmodium_spp
References
- ↑ Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, Carlton JM, Pain A, Nelson KE, Bowman S, Paulsen IT, James K, Eisen JA, Rutherford K, Salzberg SL, Craig A, Kyes S, Chan MS, Nene V, Shallom SJ, Suh B, Peterson J, Angiuoli S, Pertea M, Allen J, Selengut J, Haft D, Mather MW, Vaidya AB, Martin DM, Fairlamb AH, Fraunholz MJ, Roos DS, Ralph SA, McFadden GI, Cummings LM, Subramanian GM, Mungall C, Venter JC, Carucci DJ, Hoffman SL, Newbold C, Davis RW, Fraser CM, Barrell B. 2002. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 419:498-511