CodeOn
From CoGepedia
Contents
Coding Sequence Evolution (CodeOn)
Overview
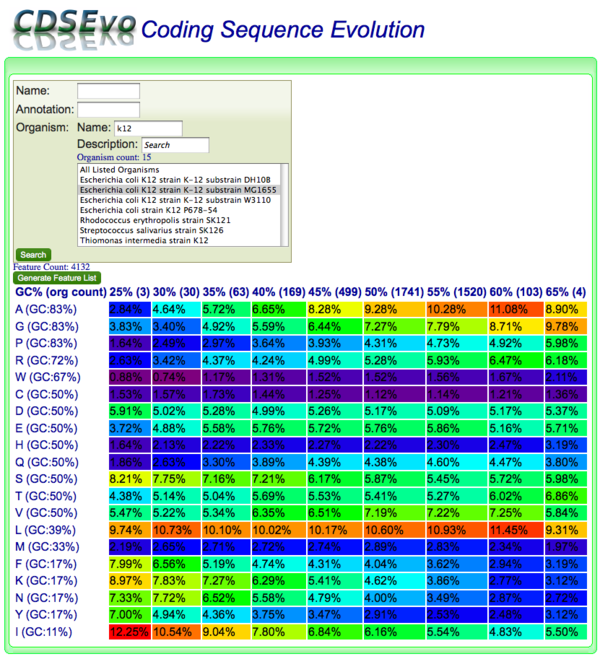
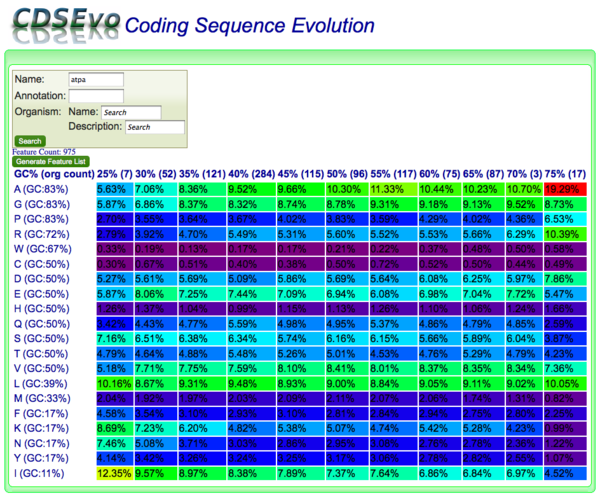
CodeOn allows you to search for CDS genomic features by name, annotation, and/or organism and display an amino acid usage table binned by the overall GC content of each CDS.
Usage
- Type in a name of a genomic feature
- Type in an annotation of a genomic feature
- Type in the name or description of an organism
Then press "search"
- When you search by genomic feature name and annotation, you can limit your search to a specific organism or set of organisms.
- If no genomic feature name or annotation is specified, but an organism or a group of organisms is, the search will get all CDSs from those organisms.
Results
The results table is organized such that:
- x-axis: CDSs are binned according to their GC content along
- y-axis: amino acids are listed according the GC content of their codons
Each cell contains:
- the relative percent usage of that amino acid for that bin
- is color-coded according to percent usage compared to all cells in the table:
- purple: lowest
- red: highest
- Order: purple < blue < green < yellow < orange < red
Linking to CodeOn
- fid= CoGe database feature id. Can have multiple specified either by fid=1&fid=2&fid=3 OR fid=1::2::3
- oid= CoGe database organism id.
- accn= Feature name
- anno= Feature annotation