Difference between revisions of "GEvo-4at-cp-vv"
| (3 intermediate revisions by the same user not shown) | |||
| Line 4: | Line 4: | ||
=Comparison of syntenic regions among ''Arabidopsis thaliana'', ''Carica papaya'', and ''Vitis vinifera''= | =Comparison of syntenic regions among ''Arabidopsis thaliana'', ''Carica papaya'', and ''Vitis vinifera''= | ||
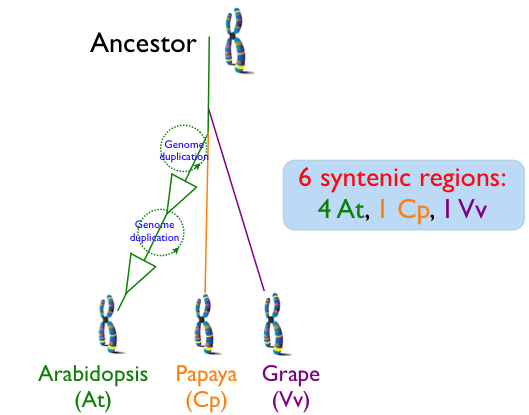
| − | Since the divergence of ''Arabidopsis thaliana'' (At), ''Carica papaya'' (Cp; papaya), and ''Vitis vinifera'' (Vv; grape), the Arabidopsis lineage has undergone two sequential whole genome duplication events, while the genomes of papaya and grape have not. This means that for every genomic regions of grape, the most basal of these lineages, there is one corresponding syntenic region in papaya, and 4 corresponding syntenic regions in arabidopsis. While whole genome duplication events contemporaneously create a copy of every chromosome and all their underlying [[Genomic Feature|genomic features]] (e.g. genes), over evolutionary time, many of the duplicated features are lost from [[homeologous]] region or its partner region. While this diploidization process will tend to return a genome's gene content to one that is more similar to the pre-[[polyploid]] ancestor, the overall structure of the genome will be changed as formerly neighboring genes may be dispersed on different derived homeologous chromosomes. This process of homeologous gene loss following whole genome duplication events is known as [[fractionation]]. Comparison of fractionated homeologs to an outgroup syntenic genomic region that has not had its own separate whole genome duplication event will yield an expected pattern: the outgroup genomic region will contain nearly the entire gene content of the fractionated syntenic regions, and the genomic arrangement of these genes will be collinear. In addition, the outgroup syntelogs of the fractionated homeologs will be intercalated with respect to one another. While fractionation may be a dominate mechanism in post-polyploid genome evolution, other mechanisms, such as gene [[transposition]] and local duplication events, are also at play that can decrease the syntenic signal by moving DNA into and out of a genomic region. | + | Since the divergence of ''Arabidopsis thaliana'' (At), ''Carica papaya'' (Cp; papaya), and ''Vitis vinifera'' (Vv; grape), the Arabidopsis lineage has undergone two sequential whole genome duplication events, while the genomes of papaya and grape have not. This means that for every genomic regions of grape, the most basal of these lineages, there is one corresponding syntenic region in papaya, and 4 corresponding syntenic regions in arabidopsis. While whole genome duplication events contemporaneously create a copy of every chromosome and all their underlying [[Genomic Feature|genomic features]] (e.g. genes), over evolutionary time, many of the duplicated features are lost from [[homeolog|homeologous]] region or its partner region. While this diploidization process will tend to return a genome's gene content to one that is more similar to the pre-[[polyploid]] ancestor, the overall structure of the genome will be changed as formerly neighboring genes may be dispersed on different derived homeologous chromosomes. This process of homeologous gene loss following whole genome duplication events is known as [[fractionation]]. Comparison of fractionated homeologs to an outgroup syntenic genomic region that has not had its own separate whole genome duplication event will yield an expected pattern: the outgroup genomic region will contain nearly the entire gene content of the fractionated syntenic regions, and the genomic arrangement of these genes will be collinear. In addition, the outgroup syntelogs of the fractionated homeologs will be intercalated with respect to one another. While fractionation may be a dominate mechanism in post-polyploid genome evolution, other mechanisms, such as gene [[transposition]] and local duplication events, are also at play that can decrease the syntenic signal by moving DNA into and out of a genomic region. |
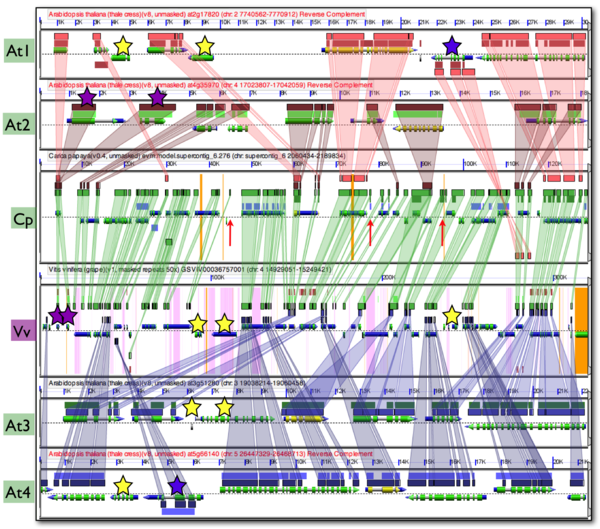
The figure shown here is a [[GEvo]] analysis of a syntenic comparison of 4 Arabidopsis regions (At1, At2, At3, Agt4) to an outgroup region of papaya (Cp), and a second outgroup region of grape (Vv). Each genomic region is represented by a panel visualizing the genomic region. The dashed line in the middle of a panel separates the top (5' left) and bottom (3' left) strand of DNA. Gene models are drawn above and below this line for each strand respectively. Gene models are colored composite arrows where the narrow gray arrow represents the extent of the gene model, blue represents mRNA, and green represents coding sequence (CDS). Other RNA genomic features, such as tRNAs, are drawn as gray arrows that are thicker than gene models (seen in At1 and At3). Regions of sequence similarity are identified by blastz and visualized as colored blocks with a separate color and track for each pair-wise comparison. Blast hits in the (++) and (+-) orientation are drawn above and below the dashed line respectively. The background of the panels are colored orange for unsequenced regions (as seen in Cp and Vv) and purple for repetitive masked sequence (as seen in Vv). | The figure shown here is a [[GEvo]] analysis of a syntenic comparison of 4 Arabidopsis regions (At1, At2, At3, Agt4) to an outgroup region of papaya (Cp), and a second outgroup region of grape (Vv). Each genomic region is represented by a panel visualizing the genomic region. The dashed line in the middle of a panel separates the top (5' left) and bottom (3' left) strand of DNA. Gene models are drawn above and below this line for each strand respectively. Gene models are colored composite arrows where the narrow gray arrow represents the extent of the gene model, blue represents mRNA, and green represents coding sequence (CDS). Other RNA genomic features, such as tRNAs, are drawn as gray arrows that are thicker than gene models (seen in At1 and At3). Regions of sequence similarity are identified by blastz and visualized as colored blocks with a separate color and track for each pair-wise comparison. Blast hits in the (++) and (+-) orientation are drawn above and below the dashed line respectively. The background of the panels are colored orange for unsequenced regions (as seen in Cp and Vv) and purple for repetitive masked sequence (as seen in Vv). | ||
| Line 11: | Line 11: | ||
These types of syntenic analyses also permit the discovery of other genomic evolution events such as: | These types of syntenic analyses also permit the discovery of other genomic evolution events such as: | ||
| − | *Inversions (Blue | + | *Inversions (Blue stars): These genes blast hits are drawn below the dashed line signifying that they are in the (+-) orientation with respect to both Cp and Vv. Since Vv is an outgroup to Cp and At, and has the same orientation as Cp, we can assume through parsimony that the inversion happened in At. |
*Gene transpositions (Yellow starts): These are genes with no similar sequence to any other genomic region, and are likely to have transposed into this region subsequent to the radiation of these lineages | *Gene transpositions (Yellow starts): These are genes with no similar sequence to any other genomic region, and are likely to have transposed into this region subsequent to the radiation of these lineages | ||
*Local gene duplications (Purple starts): These are genes that are present in two copies in At2 and Vv, and a single copy in At1 and Cp. This is seen by the wedges connecting each gene marked by a purple star to a single gene in Cp. | *Local gene duplications (Purple starts): These are genes that are present in two copies in At2 and Vv, and a single copy in At1 and Cp. This is seen by the wedges connecting each gene marked by a purple star to a single gene in Cp. | ||
| Line 17: | Line 17: | ||
In addition, comparison across multiple syntenic regions reveals annotation errors. The red arrows point to regions in Cp that have similar sequence to one or more of the other genomic regions. While there are no annotated gene models in Cp, these regions are conserved, syntenic, and are annotated as genes in their syntologous regions. | In addition, comparison across multiple syntenic regions reveals annotation errors. The red arrows point to regions in Cp that have similar sequence to one or more of the other genomic regions. While there are no annotated gene models in Cp, these regions are conserved, syntenic, and are annotated as genes in their syntologous regions. | ||
| − | This comparison can be regenerated, manipulated and modified by following this link to GEvo with these regions pre-loaded: http:// | + | This comparison can be regenerated, manipulated and modified by following this link to GEvo with these regions pre-loaded: http://genomevolution.org/r/ci31 |
Latest revision as of 14:26, 13 March 2014
Comparison of syntenic regions among Arabidopsis thaliana, Carica papaya, and Vitis vinifera
Since the divergence of Arabidopsis thaliana (At), Carica papaya (Cp; papaya), and Vitis vinifera (Vv; grape), the Arabidopsis lineage has undergone two sequential whole genome duplication events, while the genomes of papaya and grape have not. This means that for every genomic regions of grape, the most basal of these lineages, there is one corresponding syntenic region in papaya, and 4 corresponding syntenic regions in arabidopsis. While whole genome duplication events contemporaneously create a copy of every chromosome and all their underlying genomic features (e.g. genes), over evolutionary time, many of the duplicated features are lost from homeologous region or its partner region. While this diploidization process will tend to return a genome's gene content to one that is more similar to the pre-polyploid ancestor, the overall structure of the genome will be changed as formerly neighboring genes may be dispersed on different derived homeologous chromosomes. This process of homeologous gene loss following whole genome duplication events is known as fractionation. Comparison of fractionated homeologs to an outgroup syntenic genomic region that has not had its own separate whole genome duplication event will yield an expected pattern: the outgroup genomic region will contain nearly the entire gene content of the fractionated syntenic regions, and the genomic arrangement of these genes will be collinear. In addition, the outgroup syntelogs of the fractionated homeologs will be intercalated with respect to one another. While fractionation may be a dominate mechanism in post-polyploid genome evolution, other mechanisms, such as gene transposition and local duplication events, are also at play that can decrease the syntenic signal by moving DNA into and out of a genomic region.
The figure shown here is a GEvo analysis of a syntenic comparison of 4 Arabidopsis regions (At1, At2, At3, Agt4) to an outgroup region of papaya (Cp), and a second outgroup region of grape (Vv). Each genomic region is represented by a panel visualizing the genomic region. The dashed line in the middle of a panel separates the top (5' left) and bottom (3' left) strand of DNA. Gene models are drawn above and below this line for each strand respectively. Gene models are colored composite arrows where the narrow gray arrow represents the extent of the gene model, blue represents mRNA, and green represents coding sequence (CDS). Other RNA genomic features, such as tRNAs, are drawn as gray arrows that are thicker than gene models (seen in At1 and At3). Regions of sequence similarity are identified by blastz and visualized as colored blocks with a separate color and track for each pair-wise comparison. Blast hits in the (++) and (+-) orientation are drawn above and below the dashed line respectively. The background of the panels are colored orange for unsequenced regions (as seen in Cp and Vv) and purple for repetitive masked sequence (as seen in Vv).
Colored transparent wedges have been drawn connecting regions of sequence similarity. The green boxes and wedges show the typical pattern of syntenic regions. Nearly every gene in one has sequence similar to a gene in the other region which is evidence that they are orthologs. In addition, these orthologous genes have a collinear arrangement, which is evidence that these are syntenic regions being derived from the same ancestral genomic region. Arabidopsis regions At1 and At2 have wedges connecting their regions of sequence similarity to Cp while regions At3 and At4 have their regions of sequence similarity connected to Vv. This is drawn as such in order to more easily visualize synteny among all these regions, but in essence Cp and Vv are syntenic analogs for comparison to At. Each region of At has orthologs for nearly their entire gene content represented in Cp and Vv, and these orthologous genes have a collinear arrangement. As expected from the process of fractionation, the combined syntenic gene set from all the At regions are intercalated with respect to one another when compared to Cp or Vv.
These types of syntenic analyses also permit the discovery of other genomic evolution events such as:
- Inversions (Blue stars): These genes blast hits are drawn below the dashed line signifying that they are in the (+-) orientation with respect to both Cp and Vv. Since Vv is an outgroup to Cp and At, and has the same orientation as Cp, we can assume through parsimony that the inversion happened in At.
- Gene transpositions (Yellow starts): These are genes with no similar sequence to any other genomic region, and are likely to have transposed into this region subsequent to the radiation of these lineages
- Local gene duplications (Purple starts): These are genes that are present in two copies in At2 and Vv, and a single copy in At1 and Cp. This is seen by the wedges connecting each gene marked by a purple star to a single gene in Cp.
In addition, comparison across multiple syntenic regions reveals annotation errors. The red arrows point to regions in Cp that have similar sequence to one or more of the other genomic regions. While there are no annotated gene models in Cp, these regions are conserved, syntenic, and are annotated as genes in their syntologous regions.
This comparison can be regenerated, manipulated and modified by following this link to GEvo with these regions pre-loaded: http://genomevolution.org/r/ci31