CoGepedia:Current events: Difference between revisions
mNo edit summary |
Andrewnelson (talk | contribs) |
||
| (161 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
__NOTOC__ | __NOTOC__ | ||
==New Diversity Analysis Tool!== | |||
==CoGe Leadership Change== | |||
May 1, 2021 | |||
After more than a decade at the helm of CoGe, Eric Lyons is stepping down from leading CoGe. [https://btiscience.org/andrew-nelson/ Dr. Andrew Nelson] will be CoGe's new Supreme Leader. Dr. Nelson's work is deeply rooted in comparative genomics. In addition, he is a long-time member of the CoGe team and has extensive experience developing computational tools to help genomic researchers. | |||
Please welcome Dr. Nelson! | |||
[[Image:IMG 8887.jpg|600px]] | |||
(Note: Eric is still around to help answer questions along with Andrew sent to coge.genome@gmail.com and provide technical support for CoGe.) | |||
==Flash-free GEvo Update== | |||
Mar 4th, 2020 | |||
After user testing (and some bugs reported and fixed), the new Flash-free GEvo has passed. It has now been made the default GEvo. Please let us know (coge.genome@gmail.com) if you run into any problems. Also, the old one is still available by adding '-old' after GEvo in links to GEvo. | |||
Example: | |||
* '''New:''' https://genomevolution.org/coge/GEvo.pl?fid1=2717714878;fid2=2344054529;dsid2=131558 | |||
* '''Old:''' https://genomevolution.org/coge/GEvo-old.pl?fid1=2717714878;fid2=2344054529;dsid2=131558 | |||
==Flash-free GEvo Update== | |||
Feb 18th, 2020 | |||
There is a new version of GEvo available for testing: https://genomevolution.org/coge/GEvo-new.pl | |||
This new version of GEvo doesn't use Flash (which is dead or almost dead). All old GEvo links will work with the new "test" version (just add '-new' after GEvo in the URL). If you have any problems, please email us: coge.genome@gmail.com | |||
At some point in the future, the new GEvo will replace the old GEvo (and the old GEvo will be kept around as "GEvo-old.pl" | |||
==Hardware Update== | |||
Sept 16th, 2019 | |||
Our attempt to add more hard drives to CoGe's storage array failed last Fri. For some reason, the storage unit failed to recognized three different types of HDDs we tried to add. We are not sure where the problem is, and will have to bring CoGe down again to figure this out. As usual, we will post downtimes through alerts on CoGe's home page. | |||
==50,000 Genomes in CoGe== | |||
Aug 1st, 2019 | |||
CoGe passes the 50k genome mark! The majority of these are added by individual researchers using CoGe's [[LoadGenome]]. | |||
==CoGe Team Paper Accepted in Genome Biology and Evolution on Plasmodia Genome Evolution== | |||
Jan 23rd, 2019 | |||
'''Tail wags the dog? Functional gene classes driving genome-wide GC content in Plasmodium spp. ''' | |||
https://academic.oup.com/gbe/advance-article/doi/10.1093/gbe/evz015/5299488 | |||
==CoGe's Downtime explained== | |||
Nov. 2nd, 2018 | |||
On the evening of Oct. 29th, we were alerted by two researchers that CoGe was reporting failures loading new genomes. Thankfully, both of these researchers sent in the log report from CoGe. Analysis of them showed the same error: | |||
DBIx::Class::Storage::DBI::_dbh_execute(): DBI Exception: DBD::mysql::st execute failed: Duplicate entry ‘2147483647’ for key \‘PRIMARY\’ | |||
[for Statement "INSERT INTO feature ( chromosome, dataset_id, feature_type_id, start, stop, strand) VALUES ( ?, ?, ?, ?, ?, ? )" | |||
with ParamValues: 0=‘Contig16329’, 1=’129722’, 2=‘425’, 3=12001, 4=20906, 5=1] | |||
at /opt/apache2/coge/scripts/load_annotation.pl line 387 | |||
'''INT vs BIGINT''' | |||
This was bad because the Duplicate entry ‘2147483647’ was the primary key in the feature table of CoGe. This key is autogenerated and should never get duplicated. We quickly realized that the number 2147483647 was special. It is the largest value for a 4-Byte signed number. A quick check of the database structure showed that the feature table's primary key was cast as INT (a 4-Byte number) and the source of the problem was discovered. CoGe had run out of numbers. | |||
Fortunately, the fix for this is straightforward. Update the tables using this key from INT to BIGINT (https://dev.mysql.com/doc/refman/5.5/en/integer-types.html) and to an 8-Byte number. That would give a max number of 9,223,372,036,854,775,807. Which is a very, big number. | |||
'''Response Plan''' | |||
From that, we formulated a response plan: | |||
# Put CoGe into maintenance mode | |||
# Backup the database and all primary data (this runs weekly over the weekend, but we wanted to capture all the new changes). For those interested backups are made each week, stored remotely (CyVerse data store), and we keep weekly backups for four weeks and monthly backups for 4 months. | |||
# Check to see if the partition housing the CoGe database would be large enough for the updated (nope!) | |||
# Copy the database to a partition large enough to do the update (we used one of the storage partitions for experiment data that had 10TB of available space and was on SSDs for performance). | |||
# Find all tables using the feature_id (there were 5 of them) | |||
# Update those tables from INT to BIGINT for the feature_id | |||
# Check the database for other tables hitting that limit and update those for good measure | |||
# Double the tables to make sure nothing was missed | |||
# After the database was updated, either move it back to the dedicated partition or, if the partition was too small, create a new partition and then move back | |||
# Bring CoGe back up and test | |||
Another item in the response plan was to make sure we posted regular updates to on the maintenance page as well as CoGe's twitter account. Nothing is worse than needing a research tool, having it down, and having no idea when it may be back up. While we weren't sure exactly when we'd get it back up, we wanted to make sure people knew we were working on it, checking regularly, and could be contacted for whatever reason. | |||
Overall, the plan worked perfectly and CoGe was up and running again in about 4 days. The only hiccup was that access to CoGe and CoGe's servers is controlled through LDAP run by CyVerse. The night we put CoGe into maintenance mode, CyVerse's LDAP system crashed. While the CyVerse team was amazing it getting that recovered, it did stall our work for a morning (minus an hour of panic when we thought we had been locked out of CoGe for reasons unknown.) | |||
'''How long will this take?''' | |||
Unfortunately, the plan took a long time to complete. The reason for this is the shear size of CoGe's database. Even working on fast servers, SSDs, fast networks -- it takes a long time to move 2.2TB between servers. Table updates took between 12-40 hours each (they they were run in parallel). While the wait was excruciating at times, the main worry was that if any of these steps had an error, we would lose days of time. Fortunately, this didn't happen. Having a plan and a checklist was central to making sure that everything was done in the correct order, without having to restart. (e.g., if we messed up the database update, it would take a half day to pull a new copy of the database from the remote server, etc.) | |||
'''Retrospective''' | |||
The only error we made was not foreseeing that we were running out of numbers (filling the number space) for CoGe's feature table. Reviewing notes, this was identified as a future item two years ago (with the note that this would probably be a problem in two years). We didn't do the update at that time because we didn't have money to spend on SSDs for a large enough partition for the MySQl database. Well, we didn't want to spend that much money at that time knowing that in one-two years we could get twice the storage for half the cost (CoGe is a pretty lean project). | |||
However, regardless of when we did the update, the downtime would have been the same. There is no way to do this kind of an update to the database without putting CoGe into maintenance mode. Every time a researcher uses CoGe, the database has changes made (tracking analyses, updating links, etc.) and best practices is to put the database in a state where no changes are happening before updating these tables. | |||
'''But, we do want to apologize to all the researchers using CoGe.''' The one thing we could have done differently is to plan for this downtime, post an alert on CoGe giving a two week notice, and do our best to ensure that researchers could plan for this downtime as well. For that, we are very sorry. | |||
-Eric Lyons (on behalf of the CoGe Team) | |||
==Workshop at LANGEBIO CINVESTAV== | |||
Sept. 17th, 2018 | |||
Course material: [[2018 LANGEBIO CINVESTAV: Intro to genome management and analysis with CoGe]] | |||
==Publication on how to use CoGe for Plasmodium researchers== | |||
April 3rd, 2018 | |||
[https://academic.oup.com/database/article/doi/10.1093/database/bay030/4960029 A tutorial of diverse genome analysis tools found in the CoGe web-platform using Plasmodium spp. as a model] | |||
==10,000th genome loaded by researchers into CoGe== | |||
Mar. 13 2018 | |||
Sometime yesterday, the 10,000 genome was loaded into CoGe by researchers! | |||
Researchers add on average 5-6 new genomes per day, 365 days a year. | |||
[[File:Screen Shot 2018-03-13 at 11.05.25 AM.png]] | |||
==EPIC-CoGe Published in Bioinformatics== | |||
Feb. 20 2018 | |||
[https://academic.oup.com/bioinformatics/advance-article/doi/10.1093/bioinformatics/bty106/4883494 EPIC-CoGe: Managing and Analyzing Genomic Data ] | |||
Andrew D L Nelson, Asher K Haug-Baltzell, Sean Davey, Brian D Gregory, Eric Lyons | |||
Bioinformatics, bty106, https://doi.org/10.1093/bioinformatics/bty106 | |||
==LoadExp+== | |||
July 20th 2017 | |||
The new version of the LoadExperiment tool and its rebranding to [[LoadExp+]] is now complete. Read more in the July issue of Plant Direct [http://onlinelibrary.wiley.com/doi/10.1002/pld3.8/abstract here]. | |||
== GC% values added to API == | |||
July 20th 2017 | |||
when fetching a feature via the CoGe API, the percentage GC, AT, and NX values and the wobble percentage values are now included with the sequence and other feature info. | |||
==Multiple fastq support== | |||
May 9th 2017 | |||
Support for multiple fastq files when using BBDuk and BWA-MEM in LoadExp+ has been added. | |||
==Recommend Genomes for Certification== | |||
April 3rd 2017 | |||
To distinguish official genomes from copies or user-submitted data, we have added the "certified" indicator. Official genomes are manually assigned this status by members of the CoGe team. Certified genomes appear at the top of search results so they can easily be chosen when there are multiple genomes for a given species. When viewing a genome on the GenomeInfo page, users can now click a link to notify the CoGe team to recommend that they genome be certified. | |||
==NGS Analysis Performance Improvements== | |||
March 13th 2017 | |||
The NGS trimming and read-mapping pipelines in the [[LoadExperiment]] tool have been streamlined for a major performance boost. Compressed FASTQ input files (.gz, .bz2) are no longer automatically decompressed since most trimming and mapping programs support compressed inputs. Also, SAM-to-BAM conversion and BAM sorting steps are now performed on piped output from the mapping programs rather than creating intermediate files. | |||
==GATK HaplotypeCaller GVCF Now Available== | |||
March 13th 2017 | |||
The [[LoadExperiment]] tool now offers the GATK HaplotypeCaller single-sample GVCF SNP-calling option in addition to the variant-only option (per the Usage Examples described [https://software.broadinstitute.org/gatk/documentation/tooldocs/current/org_broadinstitute_gatk_tools_walkers_haplotypecaller_HaplotypeCaller.php here]). | |||
[[FIle:Gvcf.png|500px]] | |||
==BBDuk Trimmer Now Available== | |||
March 13th 2017 | |||
The [[LoadExperiment]] tool now offers the BBDuk trimmer as part of the NGS analysis pipelines. | |||
[[FIle:Bbduk.png|500px]] | |||
==Improved Search Results Interface== | |||
January 5th 2017 | |||
The "Google Drive" styled data browser featured in My Data is now also available for browsing search results. Search results, such as organisms, genomes, features, experiments, and notebooks, can be viewed, modified, shared, and sent to CoGe's analysis tools (SynMap, CoGeBlast, etc). | |||
[[FIle:search ui.png|500px]] | |||
==EPIC-CoGe Genome Browser Update== | |||
December 12th 2016 | |||
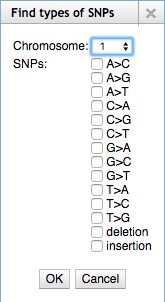
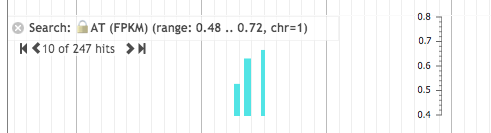
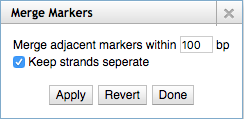
EPIC-CoGe, a genome browser built on top of [http://jbrowse.org/ JBrowse], has been updated with new data management, search and online analysis features. Users can manage CoGe experiments and notebooks from within the genome browser. Features of any type (gene, CDS, etc) can be searched for by name. Data tracks can be searched for max, min or a range of data values. SNP tracks be searched for particular types of SNPs. Tracks can be searched for data that overlaps features. Users can drag and drop tracks onto each to find overlapping data or non-overlapping data or to merge the tracks into a single track. A normalize transform has been added. Search result tracks can be converted to marker tracks and markers can be merged to remove gaps of adjustable length. Search results, transformed data and merged tracks can all be saved in CoGe as new experiments or exported to CyVerse via IRODS. Tracks can be filtered by name, metadata, tags, data type and whether or not they are private, public or shared. Support has been added for data beyond the [-1,1] range and tracks can be autoscaled on the fly. Online help has been added. | |||
[[FIle:Feature_search.png]] | |||
[[File:Range_search.png]] | |||
[[File:SNP_search.png]] | |||
[[File:search_results.png]] | |||
[[File:Combine_tracks.png|600px]] | |||
[[File:Merge_markers.png]] | |||
[[File:Filter_options.png]] | |||
==Advanced Search Features== | |||
December 12th 2016 | |||
Search results can now be filtered by various criteria available through a new GUI. Just click "advanced" next to the "Search database" input at the top of every page to access the new filters. | |||
[[File:Advanced search.png|200px]] | |||
==New Worldwide Usage Map== | |||
December 12th 2016 | |||
We've updated the site usage map on the front page to display live up-to-the-second data in a fully interactive map using Google Analytics API and Plotly. | |||
[[File:Usage_map.png|400px]] | |||
==CoGe Future Directions Survey: Tell us what you want!== | |||
November 10th 2016 | |||
There are several new features we'd like to implement in CoGe and would like your feedback to prioritize them. Please take 15 seconds to select one (or write in your own): | |||
http://goo.gl/nizYYz | |||
== BWA-MEM Aligner Added == | |||
November 8th 2016 | |||
The BWA-MEM aligner was added to the list of supported alignment programs in [[LoadExperiment]]. | |||
== New Look, Same Great Tools! == | |||
November 8th 2016 | |||
CoGe looks a little better now! We updated the user interface to a more current theme (the first update in 10 years). Please let us know of any problems, comments, or suggestions. | |||
== CoGe User-Data Association Graph == | |||
November 3rd 2016 | |||
This force directed graph shows users and the data to which they've been associated in CoGe: | |||
[[File:CoGe-user-data-small.png|500px]] | |||
* Blue nodes: Users with data, size of node corresponds to number of data sets (genomes, experiments, etc) | |||
* Purple nodes: Genomes | |||
* Green nodes: Experiments | |||
* Red nodes: Notebooks (collections) of data | |||
== FractBias Publication== | |||
October 29th 2016 | |||
FractBias has been published in Bioinformatics: | |||
http://bioinformatics.oxfordjournals.org/content/early/2016/10/26/bioinformatics.btw666.abstract | |||
==JBrowse Renewal Survey== | |||
September 23rd 2016 | |||
CoGe's genome browser is built on JBrowse. Please take time to provide feedback to the JBrowse proejct on future development. | |||
https://docs.google.com/forms/d/e/1FAIpQLSdfMxcnuP_ikEOm0HVbdCP6bXCZELQbcAJdmv-Ou2ZEEKUcIA/viewform?c=0&w=1 | |||
"This survey is aimed at users (and potential users) of GMOD genome databases, especially the JBrowse genome browser. It will directly inform the priorities for renewal of the R01 that funds JBrowse software development and the GMOD helpdesk, and your time in filling it out is GREATLY appreciated." | |||
==Favorite Stuff== | |||
September 20th 2016 | |||
You can now mark genomes, experiments, and notebooks as "favorites" (as indicated by a yellow star). Your favorites show up at the top of the list in search results, making it easier to find your preferred data. | |||
<gallery> | |||
File:Favorites.png|Favorites section in My Data | |||
File:Synmap_favorite.png|Favorite genome shown at top of SynMap search results | |||
File:Screen Shot 2016-09-22 at 1.06.37 PM.png|You can favorite a genome in GenomeInfo | |||
</gallery> | |||
==Normalization no longer required!== | |||
September 6th 2016 | |||
Normalization of quantitative experiment data is no longer required when loading new experiments. Previously, all quantitative experiment values were required to be between [0,1]. Normalization can still be optionally selected if desired. | |||
==Certified Genomes== | |||
August 13th 2016 | |||
A few years ago all of the genomes in CoGe were from NCBI, an official source. Since the addition of the LoadGenome tool users have added/copied 9000+ new genomes into the system. To distinguish official genomes from copies or user-submitted data, we have added the "certified" indicator. Official genomes are manually assigned this status by members of the CoGe team. Look for the Certified Genome icon at the end of the genome description. | |||
[[File:certified_icon.png]] | |||
Example: https://genomevolution.org/coge/GenomeInfo.pl?gid=16911 | |||
==Server Upgrade== | |||
July 13th 2016 | |||
The CoGe webserver was upgraded for performance enhancements. | |||
* A new system with 44 very fast cores, upgraded from an "antique" 32-core system | |||
* The MySQL database was mapped to a faster and more reliable storage engine (InnoDB) | |||
==New Genome Browser Features!== | |||
May 3rd 2016 | |||
Several powerful new search features were added to the CoGe JBrowse genome browser: | |||
* Search for features by name: [[Tutorials#How_to_search_for_features_in_EPIC-CoGe|Video Tutorial]] | |||
* Search quantitative tracks for max, min, or range of values: [[Tutorials#How_to_search_SNP_tracks_in_EPIC-CoGe|Video Tutorial]] | |||
* Search SNP tracks for overlap within features or for particular types of SNPs: [[Tutorials#How_to_search_SNP_tracks_in_EPIC-CoGe|Video Tutorial]] | |||
* Download data for entire tracks or regions of tracks: [[Tutorials#How_to_download_data_in_EPIC-CoGe|Video Tutorial]] | |||
Try out these new features by browsing A. thaliana: http://genomevolution.org/coge/GenomeView.pl?gid=16911 | |||
==Major Software Update== | |||
March 28th 2016 | |||
Several software components were updated resulting in major improvements, including: | |||
* the API now generates much faster responses | |||
* JBrowse upgraded to v1.12.1 with an improved Track Selector | |||
* LAST aligner upgraded to v731 for performance improvement | |||
==CoGe Workshop for Plant Reproduction 2016== | |||
March 18th 2016 | |||
Workshop information and materials: [[2016 Plant Reproduction]] | |||
==New ChIP-seq Analysis Pipeline Availablel!== | |||
March 4th 2016 | |||
A new analysis pipeline is now available (beta release) for analyzing [https://en.wikipedia.org/wiki/ChIP-sequencing chromatin immunoprecipitation sequence (ChIP-seq)]. Upload input/replicate FASTQ files and get peak results. As far as we know this is the first web-based tool of this kind! More info is available [[ChIP-seq_Analysis_Pipeline|here]]. | |||
==New Methylation Analysis Pipelines Availablel!== | |||
Feb 15th 2016 | |||
Two new methylation analysis pipelines are now available (beta release) based on Bismark and bwameth. Developed by researcher Jeff Grover and incorporated into CoGe, the pipelines allow you to upload BAM/FASTQ files and quantify methylation with the click of a button. As far as we know this is the first web-based tool of this kind! More info is available [[Methylation_Analysis_Pipeline|here]]. | |||
==New Diversity Analysis Tool Availablel!== | |||
Feb 9th 2016 | Feb 9th 2016 | ||
A new analysis pipeline is now available (beta release) for calculating basic population genetics summary statistics such as pi, theta, and Tajima's D. Upload a GCVF file for an annotated genome in CoGe and get diversity results in tabular and graphical form. As far as we know this is the first web-based tool of this kind! More info is available [[Popgen|here]]. | |||
[[ | |||
<gallery> | |||
File:popgen_example_table.png|Example table of summary statistics | |||
File:popgen_example_plot.png|Example plot of selected summary statistics | |||
</gallery> | |||
==CoGe workshop at PAG== | ==CoGe workshop at PAG== | ||
Latest revision as of 17:38, 12 May 2021
CoGe Leadership Change
May 1, 2021
After more than a decade at the helm of CoGe, Eric Lyons is stepping down from leading CoGe. Dr. Andrew Nelson will be CoGe's new Supreme Leader. Dr. Nelson's work is deeply rooted in comparative genomics. In addition, he is a long-time member of the CoGe team and has extensive experience developing computational tools to help genomic researchers.
Please welcome Dr. Nelson!
(Note: Eric is still around to help answer questions along with Andrew sent to coge.genome@gmail.com and provide technical support for CoGe.)
Flash-free GEvo Update
Mar 4th, 2020
After user testing (and some bugs reported and fixed), the new Flash-free GEvo has passed. It has now been made the default GEvo. Please let us know (coge.genome@gmail.com) if you run into any problems. Also, the old one is still available by adding '-old' after GEvo in links to GEvo.
Example:
- New: https://genomevolution.org/coge/GEvo.pl?fid1=2717714878;fid2=2344054529;dsid2=131558
- Old: https://genomevolution.org/coge/GEvo-old.pl?fid1=2717714878;fid2=2344054529;dsid2=131558
Flash-free GEvo Update
Feb 18th, 2020
There is a new version of GEvo available for testing: https://genomevolution.org/coge/GEvo-new.pl
This new version of GEvo doesn't use Flash (which is dead or almost dead). All old GEvo links will work with the new "test" version (just add '-new' after GEvo in the URL). If you have any problems, please email us: coge.genome@gmail.com
At some point in the future, the new GEvo will replace the old GEvo (and the old GEvo will be kept around as "GEvo-old.pl"
Hardware Update
Sept 16th, 2019
Our attempt to add more hard drives to CoGe's storage array failed last Fri. For some reason, the storage unit failed to recognized three different types of HDDs we tried to add. We are not sure where the problem is, and will have to bring CoGe down again to figure this out. As usual, we will post downtimes through alerts on CoGe's home page.
50,000 Genomes in CoGe
Aug 1st, 2019
CoGe passes the 50k genome mark! The majority of these are added by individual researchers using CoGe's LoadGenome.
CoGe Team Paper Accepted in Genome Biology and Evolution on Plasmodia Genome Evolution
Jan 23rd, 2019
Tail wags the dog? Functional gene classes driving genome-wide GC content in Plasmodium spp.
https://academic.oup.com/gbe/advance-article/doi/10.1093/gbe/evz015/5299488
CoGe's Downtime explained
Nov. 2nd, 2018
On the evening of Oct. 29th, we were alerted by two researchers that CoGe was reporting failures loading new genomes. Thankfully, both of these researchers sent in the log report from CoGe. Analysis of them showed the same error:
DBIx::Class::Storage::DBI::_dbh_execute(): DBI Exception: DBD::mysql::st execute failed: Duplicate entry ‘2147483647’ for key \‘PRIMARY\’ [for Statement "INSERT INTO feature ( chromosome, dataset_id, feature_type_id, start, stop, strand) VALUES ( ?, ?, ?, ?, ?, ? )" with ParamValues: 0=‘Contig16329’, 1=’129722’, 2=‘425’, 3=12001, 4=20906, 5=1] at /opt/apache2/coge/scripts/load_annotation.pl line 387
INT vs BIGINT
This was bad because the Duplicate entry ‘2147483647’ was the primary key in the feature table of CoGe. This key is autogenerated and should never get duplicated. We quickly realized that the number 2147483647 was special. It is the largest value for a 4-Byte signed number. A quick check of the database structure showed that the feature table's primary key was cast as INT (a 4-Byte number) and the source of the problem was discovered. CoGe had run out of numbers.
Fortunately, the fix for this is straightforward. Update the tables using this key from INT to BIGINT (https://dev.mysql.com/doc/refman/5.5/en/integer-types.html) and to an 8-Byte number. That would give a max number of 9,223,372,036,854,775,807. Which is a very, big number.
Response Plan
From that, we formulated a response plan:
- Put CoGe into maintenance mode
- Backup the database and all primary data (this runs weekly over the weekend, but we wanted to capture all the new changes). For those interested backups are made each week, stored remotely (CyVerse data store), and we keep weekly backups for four weeks and monthly backups for 4 months.
- Check to see if the partition housing the CoGe database would be large enough for the updated (nope!)
- Copy the database to a partition large enough to do the update (we used one of the storage partitions for experiment data that had 10TB of available space and was on SSDs for performance).
- Find all tables using the feature_id (there were 5 of them)
- Update those tables from INT to BIGINT for the feature_id
- Check the database for other tables hitting that limit and update those for good measure
- Double the tables to make sure nothing was missed
- After the database was updated, either move it back to the dedicated partition or, if the partition was too small, create a new partition and then move back
- Bring CoGe back up and test
Another item in the response plan was to make sure we posted regular updates to on the maintenance page as well as CoGe's twitter account. Nothing is worse than needing a research tool, having it down, and having no idea when it may be back up. While we weren't sure exactly when we'd get it back up, we wanted to make sure people knew we were working on it, checking regularly, and could be contacted for whatever reason.
Overall, the plan worked perfectly and CoGe was up and running again in about 4 days. The only hiccup was that access to CoGe and CoGe's servers is controlled through LDAP run by CyVerse. The night we put CoGe into maintenance mode, CyVerse's LDAP system crashed. While the CyVerse team was amazing it getting that recovered, it did stall our work for a morning (minus an hour of panic when we thought we had been locked out of CoGe for reasons unknown.)
How long will this take?
Unfortunately, the plan took a long time to complete. The reason for this is the shear size of CoGe's database. Even working on fast servers, SSDs, fast networks -- it takes a long time to move 2.2TB between servers. Table updates took between 12-40 hours each (they they were run in parallel). While the wait was excruciating at times, the main worry was that if any of these steps had an error, we would lose days of time. Fortunately, this didn't happen. Having a plan and a checklist was central to making sure that everything was done in the correct order, without having to restart. (e.g., if we messed up the database update, it would take a half day to pull a new copy of the database from the remote server, etc.)
Retrospective
The only error we made was not foreseeing that we were running out of numbers (filling the number space) for CoGe's feature table. Reviewing notes, this was identified as a future item two years ago (with the note that this would probably be a problem in two years). We didn't do the update at that time because we didn't have money to spend on SSDs for a large enough partition for the MySQl database. Well, we didn't want to spend that much money at that time knowing that in one-two years we could get twice the storage for half the cost (CoGe is a pretty lean project).
However, regardless of when we did the update, the downtime would have been the same. There is no way to do this kind of an update to the database without putting CoGe into maintenance mode. Every time a researcher uses CoGe, the database has changes made (tracking analyses, updating links, etc.) and best practices is to put the database in a state where no changes are happening before updating these tables.
But, we do want to apologize to all the researchers using CoGe. The one thing we could have done differently is to plan for this downtime, post an alert on CoGe giving a two week notice, and do our best to ensure that researchers could plan for this downtime as well. For that, we are very sorry.
-Eric Lyons (on behalf of the CoGe Team)
Workshop at LANGEBIO CINVESTAV
Sept. 17th, 2018
Course material: 2018 LANGEBIO CINVESTAV: Intro to genome management and analysis with CoGe
Publication on how to use CoGe for Plasmodium researchers
April 3rd, 2018
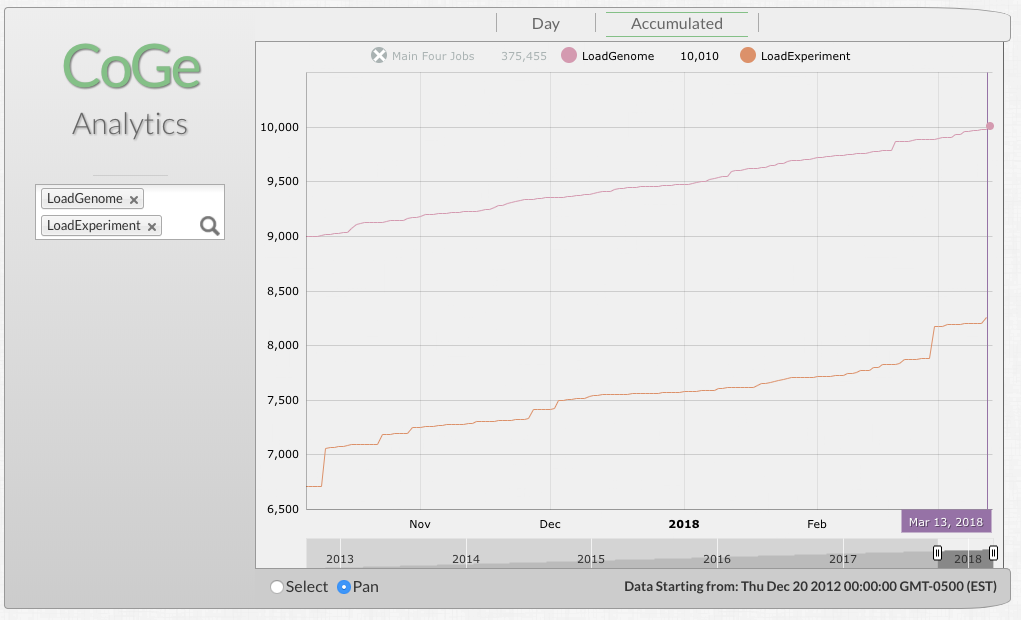
10,000th genome loaded by researchers into CoGe
Mar. 13 2018
Sometime yesterday, the 10,000 genome was loaded into CoGe by researchers!
Researchers add on average 5-6 new genomes per day, 365 days a year.
EPIC-CoGe Published in Bioinformatics
Feb. 20 2018
EPIC-CoGe: Managing and Analyzing Genomic Data
Andrew D L Nelson, Asher K Haug-Baltzell, Sean Davey, Brian D Gregory, Eric Lyons
Bioinformatics, bty106, https://doi.org/10.1093/bioinformatics/bty106
LoadExp+
July 20th 2017
The new version of the LoadExperiment tool and its rebranding to LoadExp+ is now complete. Read more in the July issue of Plant Direct here.
GC% values added to API
July 20th 2017
when fetching a feature via the CoGe API, the percentage GC, AT, and NX values and the wobble percentage values are now included with the sequence and other feature info.
Multiple fastq support
May 9th 2017
Support for multiple fastq files when using BBDuk and BWA-MEM in LoadExp+ has been added.
Recommend Genomes for Certification
April 3rd 2017
To distinguish official genomes from copies or user-submitted data, we have added the "certified" indicator. Official genomes are manually assigned this status by members of the CoGe team. Certified genomes appear at the top of search results so they can easily be chosen when there are multiple genomes for a given species. When viewing a genome on the GenomeInfo page, users can now click a link to notify the CoGe team to recommend that they genome be certified.
NGS Analysis Performance Improvements
March 13th 2017
The NGS trimming and read-mapping pipelines in the LoadExperiment tool have been streamlined for a major performance boost. Compressed FASTQ input files (.gz, .bz2) are no longer automatically decompressed since most trimming and mapping programs support compressed inputs. Also, SAM-to-BAM conversion and BAM sorting steps are now performed on piped output from the mapping programs rather than creating intermediate files.
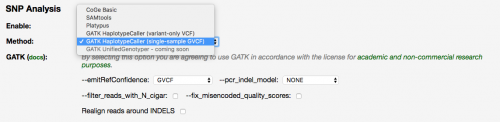
GATK HaplotypeCaller GVCF Now Available
March 13th 2017
The LoadExperiment tool now offers the GATK HaplotypeCaller single-sample GVCF SNP-calling option in addition to the variant-only option (per the Usage Examples described here).
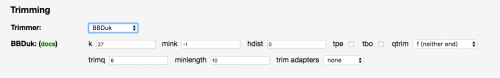
BBDuk Trimmer Now Available
March 13th 2017
The LoadExperiment tool now offers the BBDuk trimmer as part of the NGS analysis pipelines.
Improved Search Results Interface
January 5th 2017
The "Google Drive" styled data browser featured in My Data is now also available for browsing search results. Search results, such as organisms, genomes, features, experiments, and notebooks, can be viewed, modified, shared, and sent to CoGe's analysis tools (SynMap, CoGeBlast, etc).
EPIC-CoGe Genome Browser Update
December 12th 2016
EPIC-CoGe, a genome browser built on top of JBrowse, has been updated with new data management, search and online analysis features. Users can manage CoGe experiments and notebooks from within the genome browser. Features of any type (gene, CDS, etc) can be searched for by name. Data tracks can be searched for max, min or a range of data values. SNP tracks be searched for particular types of SNPs. Tracks can be searched for data that overlaps features. Users can drag and drop tracks onto each to find overlapping data or non-overlapping data or to merge the tracks into a single track. A normalize transform has been added. Search result tracks can be converted to marker tracks and markers can be merged to remove gaps of adjustable length. Search results, transformed data and merged tracks can all be saved in CoGe as new experiments or exported to CyVerse via IRODS. Tracks can be filtered by name, metadata, tags, data type and whether or not they are private, public or shared. Support has been added for data beyond the [-1,1] range and tracks can be autoscaled on the fly. Online help has been added.
Advanced Search Features
December 12th 2016
Search results can now be filtered by various criteria available through a new GUI. Just click "advanced" next to the "Search database" input at the top of every page to access the new filters.
New Worldwide Usage Map
December 12th 2016
We've updated the site usage map on the front page to display live up-to-the-second data in a fully interactive map using Google Analytics API and Plotly.
CoGe Future Directions Survey: Tell us what you want!
November 10th 2016
There are several new features we'd like to implement in CoGe and would like your feedback to prioritize them. Please take 15 seconds to select one (or write in your own):
BWA-MEM Aligner Added
November 8th 2016
The BWA-MEM aligner was added to the list of supported alignment programs in LoadExperiment.
New Look, Same Great Tools!
November 8th 2016
CoGe looks a little better now! We updated the user interface to a more current theme (the first update in 10 years). Please let us know of any problems, comments, or suggestions.
CoGe User-Data Association Graph
November 3rd 2016
This force directed graph shows users and the data to which they've been associated in CoGe:
- Blue nodes: Users with data, size of node corresponds to number of data sets (genomes, experiments, etc)
- Purple nodes: Genomes
- Green nodes: Experiments
- Red nodes: Notebooks (collections) of data
FractBias Publication
October 29th 2016
FractBias has been published in Bioinformatics:
http://bioinformatics.oxfordjournals.org/content/early/2016/10/26/bioinformatics.btw666.abstract
JBrowse Renewal Survey
September 23rd 2016
CoGe's genome browser is built on JBrowse. Please take time to provide feedback to the JBrowse proejct on future development.
"This survey is aimed at users (and potential users) of GMOD genome databases, especially the JBrowse genome browser. It will directly inform the priorities for renewal of the R01 that funds JBrowse software development and the GMOD helpdesk, and your time in filling it out is GREATLY appreciated."
Favorite Stuff
September 20th 2016
You can now mark genomes, experiments, and notebooks as "favorites" (as indicated by a yellow star). Your favorites show up at the top of the list in search results, making it easier to find your preferred data.
-
Favorites section in My Data
-
Favorite genome shown at top of SynMap search results
-
You can favorite a genome in GenomeInfo
Normalization no longer required!
September 6th 2016
Normalization of quantitative experiment data is no longer required when loading new experiments. Previously, all quantitative experiment values were required to be between [0,1]. Normalization can still be optionally selected if desired.
Certified Genomes
August 13th 2016
A few years ago all of the genomes in CoGe were from NCBI, an official source. Since the addition of the LoadGenome tool users have added/copied 9000+ new genomes into the system. To distinguish official genomes from copies or user-submitted data, we have added the "certified" indicator. Official genomes are manually assigned this status by members of the CoGe team. Look for the Certified Genome icon at the end of the genome description.
Example: https://genomevolution.org/coge/GenomeInfo.pl?gid=16911
Server Upgrade
July 13th 2016
The CoGe webserver was upgraded for performance enhancements.
- A new system with 44 very fast cores, upgraded from an "antique" 32-core system
- The MySQL database was mapped to a faster and more reliable storage engine (InnoDB)
New Genome Browser Features!
May 3rd 2016
Several powerful new search features were added to the CoGe JBrowse genome browser:
- Search for features by name: Video Tutorial
- Search quantitative tracks for max, min, or range of values: Video Tutorial
- Search SNP tracks for overlap within features or for particular types of SNPs: Video Tutorial
- Download data for entire tracks or regions of tracks: Video Tutorial
Try out these new features by browsing A. thaliana: http://genomevolution.org/coge/GenomeView.pl?gid=16911
Major Software Update
March 28th 2016
Several software components were updated resulting in major improvements, including:
- the API now generates much faster responses
- JBrowse upgraded to v1.12.1 with an improved Track Selector
- LAST aligner upgraded to v731 for performance improvement
CoGe Workshop for Plant Reproduction 2016
March 18th 2016
Workshop information and materials: 2016 Plant Reproduction
New ChIP-seq Analysis Pipeline Availablel!
March 4th 2016
A new analysis pipeline is now available (beta release) for analyzing chromatin immunoprecipitation sequence (ChIP-seq). Upload input/replicate FASTQ files and get peak results. As far as we know this is the first web-based tool of this kind! More info is available here.
New Methylation Analysis Pipelines Availablel!
Feb 15th 2016
Two new methylation analysis pipelines are now available (beta release) based on Bismark and bwameth. Developed by researcher Jeff Grover and incorporated into CoGe, the pipelines allow you to upload BAM/FASTQ files and quantify methylation with the click of a button. As far as we know this is the first web-based tool of this kind! More info is available here.
New Diversity Analysis Tool Availablel!
Feb 9th 2016
A new analysis pipeline is now available (beta release) for calculating basic population genetics summary statistics such as pi, theta, and Tajima's D. Upload a GCVF file for an annotated genome in CoGe and get diversity results in tabular and graphical form. As far as we know this is the first web-based tool of this kind! More info is available here.
-
Example table of summary statistics
-
Example plot of selected summary statistics
CoGe workshop at PAG
Jan 5th 2016
Come check out our 2-hour workshop at the Plant and Animal Genome Conference on January 9th from 4pm - 6pm!
More info here: https://pag.confex.com/pag/xxiv/webprogram/Session3102.html
CoGe workshop at the University of Chile, Santiago
Dec 18th 2015
There is an iPlant workshop held at the University of Chile with a CoGe session: details
New site header with Database Search!
Dec 7th 2015
Check out our new site header (the gray menu bar at the top). It provides a valuable new feature, Database Search, that allows you to search the entire CoGe database of organisms, genomes, and experiments. Thanks to undergraduate student worker Alex Frank for his great work on this feature!
HISAT2 aligner now available
Nov 2nd 2015
The HISAT2 aligner is now part of the RNA-seq pipelines in addition to GSNAP and Tophat.
Anyone using CoGe in Undergraduate Classes?
Sep 14th 2015
Someone asked us if there is anyone using CoGe in undergraduate classes for teaching RNASeq analyses. If you are using CoGe in undergraduate education, please let us know: coge.genome@gmail.com
CoGe Workshop at the Plant Genome Evolution Conference
Aug 26th 2015
There will be a one hour hands-on CoGe workshop at this year's Plant Genome Evolution Conference on Sept. 6th from 5:30-6:30pm
Announcement: http://www.plantgenomeevolution.com/coge-workshop.asp
Workshop's tutorials: 2015 Plant Genome Evolution Workshop
Load Genome and Annotation Updates
July 24th 2015
The user interfaces for loading genomes and annotations have been updated to a more user-friendly format. The "wizard" design guides you through the process step-by-step. See LoadGenome to try it out!
MyProfile Upgrade
June 18th 2015
The My Profile page was majorly upgraded to look better, run faster, and be easier to use:
- Added sorting based on name and date
- 5x speed-up in loading/refreshing
- New look!
Try it here (login required): https://genomevolution.org/coge/User.pl
CoGePedia Software Update
June 1st 2015
We updated CoGePedia to the latest version of MediaWiki and switched to a cleaner theme!
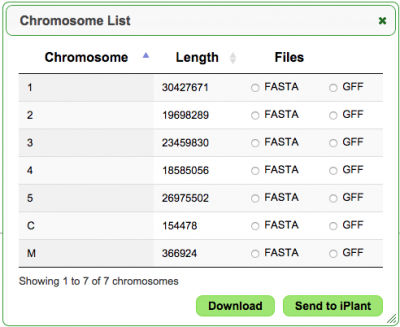
New contig browser in GenomeInfo
May 25th 2015
GenomeInfo now provides a searchable chromosome/contig table (see "List" under "Statistics") that allows the chromosome/contig sequence or annotation to be downloaded or exported to the iPlant Data Store.
Try it here: https://genomevolution.org/coge/GenomeInfo.pl?gid=16911
Unscheduled Maintenance
April 27th 2015
Our main server experienced a hardware failure last week and we had to fail over to a backup system. The motherboard was replaced and the problem was resolved. We apologize for any inconvenience due to downtime.
New SNP Pipelines Available!
March 30th 2015
A new and improved version of Load Experiment is now available. In addition to having a more user-friendly interface, the tool enables three new SNP pipelines: Platypus, SAMtools, and CoGe-basic. Just select a FASTQ file (or set of files) or a BAM file, and the tool will identify SNPs for downstream visualization in JBrowse. See LoadExperiment for more information.
Chocolate Genome Available!
January 26th 2015
We loaded the Theobroma cacao genome based on this publication: http://genomebiology.com/2013/14/6/r53#sec8
Here it is in all its deliciousness:
- Genome record: https://genomevolution.org/CoGe/GenomeInfo.pl?gid=25287
- Genome browser: https://genomevolution.org/CoGe/GenomeView.pl?gid=25287
Unscheduled Maintenance
January 21st 2015
After being moved to a new location the CoGe server crashed and was down for a day while we repaired the database. We do apologize for the inconvenience this caused, but are happy to report that everything is available and back online. Please let us know if you identify any problems.
Added support for WIG and BedGraph files
January 16th 2015
Now you can load experimental measurement data from WIG and BedGraph files in LoadExperiment.
User-contributed data is a huge success!
January 15th 2015
It has been approximately two years since CoGe added the capability for researchers to add their own genome sequence and functional/diversity genomics data to CoGe. In that time over 3000 genomes and 3500 experiments (quantitative genomic datasets) have been added by researchers from all over the world! And over 1000 new users have logged into CoGe! Thanks to this user community for making CoGe a leading place for all types of genomic analysis and visualization!
42 Methylome Datasets Published for Recently Diverged Arabidopsis thaliana linage
January 9th 2015
Recently published in PLOS Genetics: http://dx.plos.org/10.1371/journal.pgen.1004920
Century-scale Methylome Stability in a Recently Diverged Arabidopsis thaliana Lineage
Jörg Hagmann , Claude Becker , Jonas Müller, Oliver Stegle, Rhonda C. Meyer, George Wang, Korbinian Schneeberger, Joffrey Fitz, Thomas Altmann, Joy Bergelson, Karsten Borgwardt, Detlef Weigel
EPIC-CoGe used to distribute and visualize data: https://genomevolution.org/CoGe/NotebookView.pl?lid=621
CoGe at the Plant & Animal Genome Conference
January 7th 2015
There's a wonderful two-hour CoGe workshop at PAG this year entitled "Genome Management and Analysis with CoGe"!
Date: Saturday, January 10, 2015
Time: 4:00 PM-6:10 PM
Room: California
Presenters: Dr. Eric Lyons and Dr. Haibao Tang
With over 23,000 genomes from 17,000 different organisms, CoGe is where most people are managing and analyzing their genomic data. This two hour workshop will provide in-depth, hands-on training: Managing your genome in CoGe: How to add your own genome to CoGe, add structural and functional annotations, keep it private, share it with collaborators, and make it fully public (http://genomevolution.org/r/8kzz) Integrating functional and diversity data: The EPIC-CoGe addition enables researchers to integrate RNASeq, BS-Seq, ChIP-Seq, DNA resequencing data, microarrays, and any other type of quantitative genomic measurements to any genome in CoGe (http://genomevolution.org/wiki/index.php/EPIC-CoGe_Tutorial) Map NGS reads to a genome: EPIC-CoGe’s RNASeq processing and SNP discovery pipelines make it easy to transform you read data into quantitative measurements. (https://www.youtube.com/watch?v=3fNyHGB02dM) Visualizing your genomic data: The EPIC-CoGe genome browser, based on JBrowse, is instantly available for any genome and associated data loaded into CoGe (http://genomevolution.org/r/9xjy) Whole genome comparisons: Identify and classify orthologous and paralogous genes, identify whole genome duplications, use synteny to order and orient contigs and scaffolds against any reference genome to create pseudo-assemblies Micro-synteny analyses: Identify gene model errors, conserved non-coding sequences, tandem gene duplications, and changes in local genome structure Multiple whole genome synteny analyses: Given a gene in one genome, find syntenic regions across multiple genomes, even if that specific gene is not present. CoGe offers a multitude of advanced on-the-fly analyses. To accomplish these, CoGe is part of the Powered by iPlant Program and leverages iPlant’s Cyberinfrastructure for computational scalability.
Phalaenopsis equestris genome published!
December 19th 2014
The Phalaenopsis equestris genome was recently published in Nature Genetics. The horse Phalaenopsis is used extensively in horticulture, breeding new orchid cultivars, and in the cut flower industry. Additionally, this is the first CAM species to have its genome sequenced and published.
- Link to Phalaenopsis equestris genome in CoGe
50+ Avian and Crocodilian genomes published!
December 11th 2014
"The Avian Phylogenomics Consortium, led by Erich Jarvis of Howard Hughes Medical Institute at Duke University, in collaboration with the International Crocodilian Genomics Working Group, has undertaken just such a project: The international consortium has sequenced the genomes of 48 bird and 3 crocodile species.
The consortium’s first findings are published today in not one, but 28 peer-reviewed papers simultaneously released in scientific journals including Science, Genome Biology, GigaScience, and others."
- Links to the Avian genomes in CoGe: https://genomevolution.org/wiki/index.php/Bird_CoGe
- Links to the Crocodilian genomes in CoGe: https://genomevolution.org/wiki/index.php/Croc_CoGe
Brassicas: Dogs of the Plant World
December 5th 2014
Fun science video by one of CoGe's collaborators, Chris Pires:
40 New Fish Genomes now Available
November 21st 2014
Many, many new fish genomes are now available in CoGe.
See this notebook for a list: https://genomevolution.org/CoGe/NotebookView.pl?nid=890
Ongoing documentation: https://genomevolution.org/wiki/index.php/Fish_Comparative_Genomics#Data
Bug Fixes and Improved Stability
November 12th 2014
Over the last several weeks we have implemented close to three dozen bug fixes in various tools, mostly involving the User Interface. As always, please contact us with and problems or suggestions.
Tutorial for integrating genomes from JGI/Phytozome
October 17th 2014
We have published a new tutorial on how to integrate a genome from JGI/Phytozome into CoGe: How to add a genome from Phytozome or JGI
This tutorial can be easily adapted to many genome sources.
Japanese eggplant genome now available
October 17th 2014
The genome for Japanese eggplant (Solanum melongena) ([1]) is now available in CoGe.
- The eggplant (Solanum melongena L.) is one of the most important vegetable crop species in Japan as well as in other Asian, Middle and Near Eastarn, Mediterranean and African countries. Read more on Japanese eggplant
- The draft genome and transcirptome of Japanese eggplant was recently published in DNA Research.
- The genome of Japanese eggplant can be accessed within CoGe here.
New Tutorial: sending genomes to the iPlant Data Store
October 1st, 2014
This tutorial shows how fast and easy it is to send genomes from CoGe to the iPlant Data Store. From there, you can use all of iPlant's tools and services for additional analyses.
Coming soon: New SNP-finding Pipelines!
September 22nd 2014
In the next few weeks we will be deploying new SNP pipelines, including the common SAMtools/Bcftools method described here.
Export data via Web Services API
September 15th 2014
In addition to a dozen or so minor bug fixes and improvements, we just added the ability to export genome sequence, annotation, and experimental data to our public web services API. Now you can send data to the iPlant Data Store or retrieve it over HTTP even quicker and easier than before. For further details, including the API specification document, see the API article.
Two Minor Improvements
September 2nd 2014
This week on the CoGe site two minor improvements were added to LoadGenome:
- Importing a file or directory from the iPlant Data Store that has spaces in its name is now supported.
- Compressed tar archives (.tgz or .tar.gz) are now supported.
Improved SynMap Search Usability
August 25th 2014
Searching for organisms using the SynMap search bar is now more powerful and easier to use:
- can search an organism by both its name and taxonomy in the same search
- can search with multiple keywords, rather than an exact phrase
New CoGe Blast Feature: Top Hits
August 17th 2014
You can now generate a list of "top hits" for selected hits in the results table. Just select some or all hits in the table and use the "Top Hits" send-to function in the dropdown menu at the bottom (see screenshot). A list of the highest-scoring results (by HSP score) are displayed in a new window according to query name.

Bug Fixes
August 17th 2014
Over the last couple weeks the CoGe Team fixed a couple dozen minor bugs in various tools within the system.
We always welcome and appreciate bug reports and general feedback from our users! Email us!
SynMap Improvement
August 12th 2014
SynMap's organism search interface is now less cluttered and easier to use, check it out: https://genomevolution.org/CoGe/SynMap.pl
CoGeBlast Improvement
August 12th 2014
CoGeBlast now displays much more detailed progress information, check it out: https://genomevolution.org/CoGe/CoGeBlast.pl?dsgid=16911?fid=1
User Interface Improvements
July 29th 2014
Over the last few months the CoGe Team has been working with experts from the lab of Angus Forbes to evaluate and improve CoGe's entire User Interface. We have conducted about 10 in-person interviews with users to gather data on how they use the system and what's not working. Based on this feedback changes have been made to almost every page in CoGe, including major improvements to the User Profile. Let us know what you think!
Tef genome now in CoGe
July 28th 2014
The genome for Tef (Eragrostis tef) is now available in CoGe.
- Tef is a millet grown as an important agricultural crop in Ethiopia and used as source of specialty foods in India, Australia and the United States. Read more on Tef
- The draft genome and transcirptome of Tef was recently published in BMC Genomics.
- The genome of Tef can be accessed within CoGe here.
User Interface Redesign Coming Soon
July 18th 2014
Over the last few months the CoGe Team has been working with experts from the lab of Angus Forbes to evaluate and improve CoGe's entire User Interface. We have conducted about 10 in-person interviews with users to gather data on how they use the system and what's not working. Next week we will unveil the improvements, stay tuned!
User Profile Improvements
July 11th 2014
The improved "Analyses" section of the User Profile now provides the ability to "favorite" and add comments to analyses. Also, a new section "Data loads" displays the real-time status of data loading events from LoadGenome, LoadExperiment, etc.

CoGe Release 5.6
June 26th 2014
We recently deployed CoGe release 5.6 which contains major improvements to job distribution, management, and reliability using our Job Execution Framework (JEX). Processes that were previously run as child processes of the web server, such as genome and quant data loading, are now more reliably run in JEX.
CoGe Security Update
June 6th 2014
All web-traffic to and from CoGe is now secured with SSL/HTTPS (heartbleed free!).
Load Experiment Web Service
May 22nd 2014
Getting large amounts of data into CoGe just got easier with the release of the Experiment Add web service available via CoGe's RESTful API.
See the API page for more information and an example of how to load an experimental data set.
SNP-finding Pipeline Available
May 20th 2014
CoGe now provides an integrated SNP-finding pipeline! Load a BAM file using LoadExperiment view an existing BAM experiment using ExperimentView and select the "Find SNPs" option. It will analyze the BAM file to identify SNPs and load them as a new experiment. For more information see Identifying_SNPs.
-
ExperimentView's "Find SNPs" function (highlighted in red)
-
SNP track (highlighted in red)
Improved Quant Track
May 9th 2014
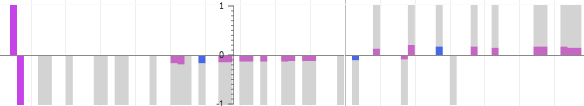
A minor improvement to the quantitative experiment track in the EPIC-CoGe viewer allows you to differentiate zero-value data points from missing data. In the track menu select the "Show background" option to draw a gray background behind data points.
-
Track menu
-
Visualization with background
Add Labels to Your Data
Apr. 18th 2014
A minor improvement to the quantitative experiment track in the EPIC-CoGe viewer allows you to add labels to your data points using the "name" field in a BED file.
CoGe Web Services API Now Available
Apr. 14th 2014
CoGe web services are now available! Search and query organisms, genomes, and data sets within our database.
Documentation:
http://docs.google.com/document/d/1GXOPIVvyTwoGR2IRjDNmdd2nj_6d8db7jx9hG8RxTDc
See the API page for more info and examples ...
RNASeq pipeline now supports TopHat/Bowtie2
Apr. 11th 2014
In addition to GSNAP, you can now select TopHat/Bowtie2 for aligning FASTQ reads to a reference genome.
See Expression Analysis Pipeline for more info.
CoGe Web Services API Under Development
Apr. 4th 2014
We are currently developing public web services for accessing and loading data in CoGe!
Here is our draft API specification document:
http://docs.google.com/document/d/1GXOPIVvyTwoGR2IRjDNmdd2nj_6d8db7jx9hG8RxTDc
See the API page for more info and examples ...
RNASeq processing pipeline now available
Apr. 3rd 2014
Have RNASeq data that you want to map to a genome in CoGe? Now you can do that quickly and easily!
- Pipeline information: Expression Analysis Pipeline
- Thanks to James Schnable for working with us to integrate his pipeline from qTeller
Awesome new dotplot viewer unveiled!
Mar. 21st 2014
We are very excited to unveil a prototype of the new CoGe dotplot viewer! SynMap currently displays a static image for the dotplot result. This new dotplot viewer employs the latest HTML5 browser technology to provide a fully dynamic and interactive display.
Here's an example in CoGe SynMap
Here's a generic demo:
- Single: http://genomevolution.org/jdot/test/tester.html
- Multi: http://genomevolution.org/jdot/test/tester2.html
We are distributing this code as a standalone project so that anyone can use it. It also works for applications beyond dotplot as a generic scatter plot viewer.
- http://github.com/LyonsLab/jdot
- note: this is early alpha code
Links between BAR Expressolog Tree and CoGe
Mar. 2nd 2014
Rohan Patel from Nicholas Provart's group at the University of Toronto has linked Expressolog data to CoGe for microsynteny analysis. This combination of tools lets users quickly identify common patterns of gene expression from a phylogenetic perspective and validate synteny among genomes with CoGe's tool GEvo.

Improvements to qTeller integration and GenomeView
Feb. 28th 2014
The new expression analysis pipeline now additionally generates a raw read depth track along with the previous alignment and FPKM tracks.
- Details of the pipeline: Processing RNA seq data
- Command line and arguments used in the pipeline: Expression Analysis Pipeline
Also, now you can change track color for individual experiment tracks.
Beta Release: Automated expression analysis and qTeller integration
Feb. 21st 2014
It is now possible to quickly and easily generate gene expression measurements qTeller style! Just use LoadExperiment to load a FASTQ file onto an annotated genome and our pipeline will automatically map the reads and run cufflinks. View both outputs in our JBrowse-based viewer.
Update: Integration with qTeller for automated expression analysis
Feb. 14th 2014
This week we have been testing and improving upon the expression analysis pipeline. We are in the process of finalizing the pipeline. We would like to thank James Schnable for visiting us this week.
Coming soon: Integration with qTeller for automated expression analysis
Feb. 7th 2014
This week we have been working on adding a qTeller-based expression analysis pipeline to CoGe. We should have it finished within the next week. Thanks to James Schnable for the help!
More new features to GenomeView and GenomeInfo
Jan. 31st 2014
- Change experiment track height in GenomeView
- Detailed sequence and feature statistics in GenomeInfo
- %GC content and histograms in GenomeInfo
-
Change all track heights
-
Adjusting an individual track height
-
Histograms now in GenomeInfo
Data Export to iPlant Data Store
Jan. 24th 2014
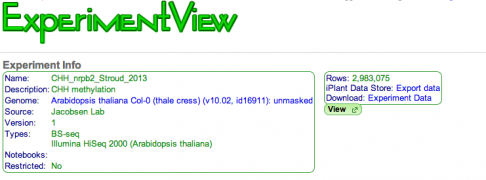
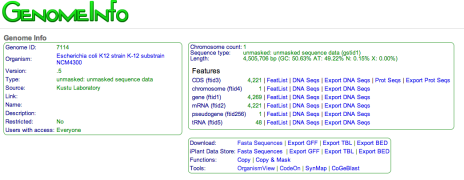
New features in ExperimentView and GenomeInfo are now available that allows you to export your experiment and genome data to the iPlant Data Store or your local system.
-
ExperimentView
-
GenomeInfo
Post-PAG updates to CoGe
Jan. 16th 2014
With PAG finished, the CoGe team has rolled-out the following new features:
- New homepage!
- Exporting and downloading experiment data from ExperimentView to your iPlant Data Store or desktop
- This feature exports the raw data file as well as the FastBit indexed database
- Metadata markup for genomes available through GenomeInfo (including pictures and links)
- Many updates to GenomeView's JBrowse
- Updated to the latest JBrowse version 1.11.1 which provides a coverage histogram view of BAM read-mapping data
- New quantitative data transformations
- Differences between experiments
- Log10 transformations of values
- Ability to save changes to quantitative track colors
- Ability to export gene models to FeatList for downstream data extraction and analysis
-
BAM coverage histogram
-
Transformations
-
Send gene models to FeatList
We had a great time at PAG this year seeing old friends and meeting new people. Please keep sending us your suggestions and improvements!
Load genome, annotation, expression, diversity, and read-mapping data in 3 minutes
Jan. 7th 2014
This video shows using EPIC-CoGe to load a genome, annotations, and lots of data for visualization in JBrowse in under three minutes (the extra 2.5 minutes are to explain the setup and validate the results): http://youtu.be/JPZ8IrPnh_8
For more info and instructions: How to load a private genome into CoGe
CoGe @ PAG
Jan. 3rd 2014
Some of the team will be at PAG this year. You can learn of our latest additions and improvements to CoGe at:
- Computer Demos: https://pag.confex.com/pag/xxii/webprogram/Paper10615.html
- EPIC: the Plant Epigenome Project: http://pag14.mapyourshow.com/5_0/sessions/sessiondetails.cfm?ScheduledSessionID=1DAC
- Banana Genomes: http://pag14.mapyourshow.com/5_0/sessions/sessiondetails.cfm?ScheduledSessionID=18AC
Happy New Year
Jan. 1st 2014
2013 brought many improvements to CoGe:
- Advanced data management
- Loading your own genomes
- EPIC-CoGe
- Integration of JBrowse
- Support for functional and diversity genomic data sets
- Lots of new genomes
Over the break, the CoGe team finally hunted down and fixed an outstanding issue with CoGe's database. We had noticed a long delay (upwards of an hour) during database inserts. At times, this would crash the database. The source of the problem was having a much too large query_cache for our database. While this would improve performance for many queries, each database insert required the query_cache to be flushed, which takes a long time when set to 256G. Lesson learned. Now, while our database performance is still quite good (4000+ queries per second), our database is again rock-solid during many inserts (e.g. loading genome annotations).
We are looking forward to 2014! As always, let us know if you have any ideas for CoGe.
-The CoGe Team: Matt, Evan, and Eric!
Major Maintenance Completed
Dec. 21st 2013
We just finished running a major maintenance on our database. We apologize if you experienced any problems or outages, particularly with LoadGenome, over the last few days! The work done should greatly improve CoGe's reliability and scalability in the future.
Upgrade EPIC-CoGe Browser to latest JBrowse version 1.10.10
Nov. 27th 2013
This release of JBrowse has lots of minor bug fixes, new features, etc. Read more about this JBrowse release: http://jbrowse.org/jbrowse-1-10-10/
- This release includes an updated sequence track with 6-frame translations. Ooh shiny!

GenBank Accession genome loader now available
Nov. 15th 2013
Genomes can now be added to CoGe by GenBank Accession in LoadGenome.
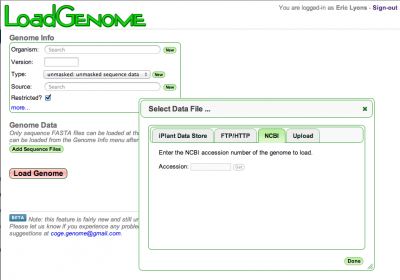
CoGe Weekly update
Nov. 8th 2013
- GenBank genome loader has been updated:
- New NCBI bacterial genomes have been loaded (~1000 new genomes): ftp://ftp.ncbi.nlm.nih.gov/genomes/GENOME_REPORTS/prokaryotes.txt
- New NCVI viral genomes have been loaded (~1000 new genomes): ftp://ftp.ncbi.nlm.nih.gov/genomes/GENOME_REPORTS/viruses.txt
- New load script will be made available through LoadGenome soon
- New feature in GenomeView:
- View Wobble GC% in Feature track
- Medicago V4 (from JCVI) is now in CoGe:
- Unmasked: http://genomevolution.org/CoGe/OrganismView.pl?dsgid=22582
- WindowMasker: http://genomevolution.org/CoGe/OrganismView.pl?dsgid=22583
- Syntenic dotplot of v4 vs. v3.5.4 of Medicago: http://genomevolution.org/r/a0da
CoGe Genome Management Updates
Oct. 31st, 2013
This past month has seen a variety of updates to CoGe including:
- Migration to new servers
- Lots of bug fixes
- Usability changes to some tools (CoGeBlast, SynMap)
- Genome Export to a user's iPlant Data Store through GenomeInfo pages
- Ability to copy and mask genomes through GenomeInfo pages
- More stability to the Job Execution Engine!
Kiwifruit genome added to CoGe
Oct. 20th, 2013
The Kiwifruit genome has been added to CoGe!
- Access the unmasked and masked version of its genome: http://genomevolution.org/CoGe/OrganismView.pl?oid=38535
- SynMap analysis of kiwi vs. grape showing kiwi has undergone two WGD since the divergence with grape (as reported): http://genomevolution.org/r/9xtx
Other resources:
- Nature paper: http://www.nature.com/ncomms/2013/131018/ncomms3640/full/ncomms3640.html
- Kiwifruit Genome Database: http://bioinfo.bti.cornell.edu/cgi-bin/kiwi/home.cgi
CoGe Server maintenance
Oct. 5th, 2013
In the process of migrating servers, doing some maintenance, testing our backups. Looks like we are back in action! (Or, at least the wiki is. . .)
UPDATE: Migration went smoothly overall. Took another week to iron out all the small bugs and configuration issues that crept up once the server was in a production environment.
iPlant is renewed!
Sep 17th, 2013
Since CoGe leverages iPlant's Cyberinfrastructure for its computational scalability, this is very good news:
BAM visualization comes to EPIC-CoGe
Sep 16th, 2013
You can now upload BAM files in Load Experiment and visualize them in CoGe. We are still updating the CoGePedia documentation for this feature, stay tuned ...
Improvements to User Profile
Sep 6th, 2013
We've added a couple new improvements to the User Profile page.
- The "Groups" section allows you to view and edit your User Groups.
- The "Analyses" section allows you to see your current and past analyses. Running jobs can canceled.
SNP visualization comes to EPIC-CoGe
July 29th, 2013
You can now upload SNP data in VCF format and visualize them in CoGe. We have also loaded 340+ resequenced Arabidopsis genomes available from the 1001 Arabidopsis project. These resequenced lines were specifically part of the MPICWang2013 project.
New CoGe Version (5.6) has moved to production
July 13th, 2013
We have deployed a new version of CoGe. Changes include:
- The EPIC-CoGe browser has many new improvements (beta-testing SNP visualization).
- Change to new version of python
- Testing CoGe's new Job Execution Framework.
We thank everyone for identifying bugs and reporting them. We are working to get them fixed as soon as possible.
EPIC-CoGe Browser has been announced on at EPIC
June 16th, 2013

The Epigenomics of Plants International Consortium has announced that the EPIC-CoGe Browser is available for testing on their homepage: https://www.plant-epigenome.org.
EPIC-CoGe Browser is ready for beta testing
June 10th, 2013
We are very happy to announce that the EPIC-CoGe Browser is ready for some early testing. Please see this EPIC-CoGe Tutorial to get started or follow this link to the EPIC-CoGe Browser for Arabidopsis thaliana.
Please email us or post to our forums any comments or questions you have.
Also, if you would like to see our requirements document: you can find it here!
New Features in EPIC-CoGe browser
June 7th, 2013
We have been working hard on improving the interface and functionality of the new EPIC-CoGe genome browser (see here).
- Drag & drop organization of experiments into notebooks
- "CoGe-like" gene annotations
- New GC-content track
- "Add all" and "Clear all" filtered tracks
Video Demonstration of EPIC-CoGe Browser
May 30th, 2013
New Feature: EPIC-CoGe Browser
April 26th, 2013
We are very excited to announce that we are moving to incorporate the amazing JBrowse viewer into CoGe's GenomeView feature. Click here to see a demo of JBrowse displaying a genome with lots of quantitative data sets. This feature is still under development (translation: there are still some major bugs), but in the coming month we will be working out the kinks and adding new features.
New Feature: LoadAnnotation
April 2nd, 2013
Now you can load your own annotation data in addition to genomic sequence and quantitative data! Click on a genome you've loaded in your User Profile page and select Load Annotation.
New Feature: NCBI Taxonomy Browser
March 8th, 2013
The LoadGenome feature now has an improved way to specify a new organism in the system. The new NCBI taxonomy browser allows you to search for the NCBI name and description for an organism and modify it if needed.
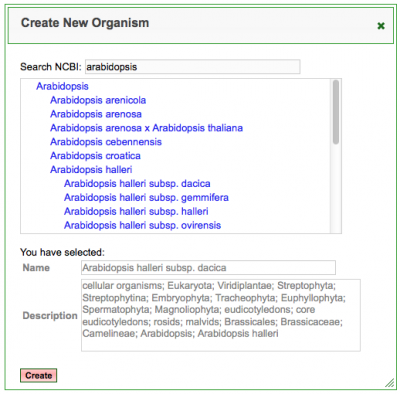
CoGe v5.5 Upgrade
Feb. 8th, 2013
Many further improvements and bug fixes have been made to the data management features in CoGe for loading and share private genomes. This upgrade marks a major change in the system that allows you to share your data with individual users as well as user groups. If you haven't seen it yet, check out the new user profile page (login required) that allows you to load and share your own genome sequence and quantitative data in CoGe. We have some ideas on further improving these features and will be working on them over the next few weeks.
CoGe at PAG!
Jan 12th, 2013
We presented a computer demonstration of the new User Data Management features in CoGe at the Plant and Animal Genome conference today. Here are the slides.
More news on EPIC-CoGe
Jan. 9th, 2013
Happy new year! Development continues on the EPIC-CoGe Browser. An overview of the project and information on progress will be posted: here.
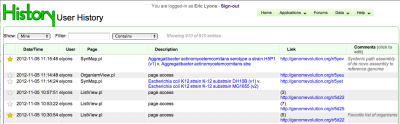
History of analyses and pages available to registered users
Nov. 5th, 2012
Improvements are continuing to CoGe's User System. If you are using a user account with CoGe , CoGe will record which pages and analyses you have run automatically. Some tools, such as SynMap, will automatically mark up those analyses in History with additional information (such as the genomes compared). If you want to revisit an analysis or page, but have lost track of it (or closed the browser tab containing it), you can retrieve it by clicking on "Data"->"History" from the menu bar in the top-right of any page.
Also, you can comment and mark specific analyses to save (and then find) for later.
CoGeBlast Update
Oct. 15th, 2012
The CoGeBlast user interface was revamped for a cleaner appearance and simpler use. The functionality remains unchanged, except the addition of a button to import target genomes from existing lists.
CoGe "Data Tab"
Oct. 5th, 2012
With the roll-out of CoGe v5 comes the ability for users to more easily organize and share their data of interest. Most of these features are found in the "Data" tab in the CoGe menu located in the upper right part of the screen:

- User Profile: Shows what information CoGe's stores about you (user name, real name, email address) and a list of your groups
- User Groups: Groups of users to which you have access. These groups are used to share lists of data
- Data Lists: Lists of data (genomes, features, experiments) to which you have access. May be private or public data. It is through these lists and User Groups that allows you to share private data with collaborators
- History: CoGe has always generated tiny links for your analysis and views of data. These are now stored for you so you may more easily find a previously run analysis.
CoGe v5 Deployment Process
Sept 24th, 2012
- 9am: We shut down the website at 9am and started the final backup and freezing of existing data and analyses.
- 10am: Database is being replicated to the iPlant Data Store and copied to a backup server for processing and conversion to new database scheme.
- 11am: All web-code and libraries were backed up and new code deployed
- 12pm: updating database
- 1pm: copying database back to iRODS
- 2pm: copying database to coge server
- 3pm: reconfiguring the system
- 3:30pm: turn on web server
- 3:31pm: Nothing works
- 3:32pm: Start debugging
- 3:34pm: Get things working -- CoGe starts!
- 4:30pm: Most major problems found and corrected
CoGe v5 Deployment
Sept 21st, 2012
CoGe v5 is planned for deployment on Sept. 24th. This new version of CoGe represents a massive revamping and extension of the user-data management system.
Key features include:
- Limited support for experimental data
- This new system is funded (in part) by a grant from the Gordon and Betty Moore Foundation to add visualization support for epigenetics data for Arabidopsis. This is known as the EPIC-CoGe project: http://www.iplantcollaborative.org/learn/news/2012/05/24/iplant-ci-leveraged-development-epic-coge-browser
- Ability to make lists and collections of data
- Lists of experiments
- Lists of genomes
- Lists of features
- Lists of lists
- Enhancements for managing and sharing private data in CoGe
- Logging user history so it is easier to find old analyses
A key part to the migration to the new version of CoGe is preserving current private data in the system and assigning them to the appropriate owner. (We have done some major changes to the underlying metadata storage database for CoGe). Please let us know if you have lost access to your data and we will get that corrected right away.
New features will be added that further integrate user specified lists into various tools in CoGe. E.g. auto-selecting a list of genomes for use in CoGeBlast instead of manually searching for all the genomes.
Many thanks to CoGe Developer Matt Bomhoff for all the work on this new version.
Please post any comments, suggestion, questions to CoGe's Forums (hosted by iPlant): https://forums.iplantcollaborative.org/viewforum.php?f=10
Phaseolus vulgaris (common bean) v1 added to CoGe
Aug 23, 2012
Released from JGI/Phytozome, it is v1 of the common bean: http://genomevolution.org/CoGe/OrganismView.pl?oid=36223
Syntenic dotplots between it and soybean (Glycine max), Phaseolus vulgaris v. Glycine max, clearly show that Phaseolus lacks the most recent Whole genome duplication in the Glycine lineage.
CoGe Paper published: Unleashing the genome of Brassica rapa
July 31th, 2012
This Open Access paper provides a set of examples of how to analyze and compare the genome of Brassica rapa. Very useful for people wanting to learn how to use CoGe or how to maximize their use of the genome of Brassica rapa:
Open Access article in Frontiers of Plant Genetics and Genomics: http://www.frontiersin.org/plant_genetics_and_genomics/10.3389/fpls.2012.00172/abstract
Also located in CoGe Tutorials sections.
Banana genome published
July 12th, 2012
The banana genome was published today in Nature: http://www.nature.com/nature/journal/vaop/ncurrent/full/nature11241.html
CoGe was used in some of the analyses (in supplementary figures), and the genome is now publicly available: http://genomevolution.org/coge/OrganismView.pl?oid=38351
Banana represents the first non-grass monocot genome to be sequenced and sheds light in the evolutionary history of the lineage as a whole.
My opinion is that the timing, placement, and make-up of the early monocot duplication events are still an open question. Some work points to an additional polyploidy event in the Poales lineage (See: Tang. et al. Angiosperm genome comparisons reveal early polyploidy in the monocot lineage. PNAS 2010; http://www.pnas.org/content/107/1/472.full). Banana, with its own series of independent series of whole genome duplications, is not the best suited for unravelling this earlier events, but these events open many avenues for additional research into the evolution and architecture of plant genomes. It will be exciting to see what similarities and differences exist between the monocots and the dicots.
Additional news pieces on banana:

Tomato genome published; the solanum hexaploidy investigated with CoGe
May 31st, 2012
The tomato genome was published in Nature earlier this week: http://www.nature.com/nature/journal/v485/n7400/
However, the current version of the tomato genome has been in CoGe for the past year (thanks to an early release of the data from the tomato genome consortium).
I've received a couple of emails inquiring about the Solanum specific hexaploidy, and this has been investigated with Haibao Tang. Overall, these analyses support that the majority of the genome is derived from a tetraploidy, but there is evidence of some regions being triplicated (perhaps through a hexaploidy).
These analyses are available: Tomato genome
Please send us your thoughts or post them on the CoGe Forum:
May 29th, 2012
If you want to load a private genome into CoGe, you need to send that genome to the CoGe team. This method makes it very easy for us to download your genome quickly!
Dedicate CoGe Forum hosted at iPlant (part of the powered by iPlant program)
May 25th, 2012
iPlant has set up a dedicated forum for CoGe: https://forums.iplantcollaborative.org/viewforum.php?f=10
Please post any CoGe questions you have to here.
EPIC-CoGe Browser
May 24th 2012
News article about this project at iPlant: http://www.iplantcollaborative.org/learn/news/2012/05/24/iplant-ci-leveraged-development-epic-coge-browser
Overview of the Epic-CoGe Browser prototype system:
Try it: http://genomevolution.org/CoGe/GenomeView.pl?z=6&x=20000&dsgid=7043&chr=1
WARNING: performance is a known issue! Some tiles in the browser may take a while to render (but are then cached).
2000 new genomes in CoGe
Apr. 30th 2012
The NCBI genome loader program was updated and run over the weekend. This resulting in about 2000 new genomes being loaded into CoGe.
Unscheduled CoGe downtime
Apr. 25th 2012
CoGe was down/offline yesterday for two reasons:
- One of iPlant's VMs was compromised and UITS (UA's IT group) shut off one of iPlant's subnets, which CoGe happens to use. This was due to a VM administered by a group collaborating with iPlant and not due to iPlant
- Since CoGe was offline, when it came up, we decided to keep it offline for a while longer in order to updated the apache web server. After apache was updated, CoGe was brought online. Unfortunately, UITS detected a security vulnerability in the SSL implementation in the new update and shut CoGe off. This last part happened at the end of the day and we weren't able to coordinate with UITS to push a fix until this morning.
While the CoGe team tries to keep as much uptime as possible, this type of downtime does happen once and a while. Our apologies to everyone whose work was interrupted or delayed due to this.
The algorithm, Last, added to SynMap
Mar. 28th 2012
Last (http://last.cbrc.jp/) has been added as a comparison algorithm in SynMap. Its performance is phenomenal! This is still under testing, so please let us know if you have any problems with it. Also, special thanks to Haibao Tang for writing the parallelized adapter for Last that is used by SynMap. Without this program, the integration would not have happened as quickly, easily, or smoothly.
CoGe used to decode the secret message in JCVI Synthetic Genome
Mar. 24th 2012
I heard that there was a secret message in the JCVI synthetic genome: Mycoplasma mycoides JCVI-syn1.0. Using CoGe, the DNA containing the secret messages was identified and decoded. Here is the walk-through of how this was done: Mycoplasma mycoides JCVI-syn1.0 Decoded.
- WARNING: contains spoilers!
- Note: this puzzle is nearly 2 years old.
For those interested in doing the puzzle, this article has a good summary of the challenge:
- Article from Sigularity Hub on the secret message: http://singularityhub.com/2010/05/24/venters-newest-synthetic-bacteria-has-secret-messages-coded-in-its-dna/
And you will probably need the original article (and the Supplementary Data):
- Original Science Article on the genome: http://www.sciencemag.org/content/329/5987/52.abstract
CoGe Forums
Mar. 2nd 2012
iPlant has a forums site available: http://forums.iplantcollaborative.org
CoGe, being part of the "Powered by iPlant" program, has a section on there for users to post questions about how to do various tasks, about CoGe in general, and provide suggestions. I'll be posting questions that are emailed to me there, but this will hopefully be a good place for people to ask questions, find answers, and help one another.
Powered by iPlant Forum: https://forums.iplantcollaborative.org/viewforum.php?f=8
The CoGe Forum: https://forums.iplantcollaborative.org/viewforum.php?f=10
BlastN Bug
Feb. 11th 2012
Mike Freeling from UC Berkeley has found an interesting bug in BlastN where a relatively large blast hit (HSP) appears/disappears depending on the amount of sequence compared between Arabidopsis and Brassica. James Schnable from UC Berkeley further characterized this by identifying a comparison that differs in 1 nucleotide (over ~750) that causes this effect. You can see images of this blast error, characterization of the blast, an breakdown of parameters used here: GEvo Blastn Bug
CoGe Server Migration
Feb. 4th 2012
CoGe's entire system has been migrated to the new server hosted by the [iplantcollaborative.org iPlant Collaborative]. This include
- A new version of CoGe (v4) that includes:
- Migration of CoGePedia to new server
- Migration of CoGe's tiny-url service (used to construct URLs that can be used to regenerated analysis -- mainly by GEvo and SynMap)
- Update to DNS for:
Please contact us if you come across any problems!
Exciting new plant genomes in CoGe
Feb. 3rd 2012
Update on genomes available from Phytozome.
The genomes of
- common bean (a crucial staple food of grad students everywhere)
- capsella (the close relative of arabidopsis, not the song by I:Scintilla)
- Syntenic dotplot: Capsella rubella - Arabidopsis lyrata
- Linum usitatissimum (common flax; linseed): http://genomevolution.org/CoGe/OrganismView.pl?oid=36226
- Syntenic dotplot: Flax -Poplar
- Gossypium raimondii (cotton): http://genomevolution.org/CoGe/OrganismView.pl?oid=36239
have both been added to iPlant CoGe. Head over and check them out. <-- But remember these genomes are protected by Fort Lauderdale for the next twelve months or until you see the genome paper.
Are we missing plant genomes you'd like to be studying? Let us know!.
iPlant User Management System Update
Dec. 18th 2011
The Data security model of CoGe has been updated. This includes creating CoGe Groups which permits the creation of user groups. These user groups may access a private set of genomes that is not accessible to other users of CoGe.
To use this, you will need to create an account with iPlant in order to be a registered CoGe user:
Major CoGe Update (version 4)
Dec. 4th 2011
Work is nearing completion for a new version of CoGe. While there are many minor improvements, additions, and changes to the tools, the major improvements are on the backend of the system including:
- New server hosted by iPlant: This means that the primary CoGe server will be located at the University of Arizona, Tucson
- Vastly expanded storage to hold even more genomes
- Enables the storage of metagenomes (as those datasets can be quite large)
- Modularized installation and centralized configuration: permits the rapid deployment of custom versions of CoGe (for those that may want a version of CoGe specific to their group of organisms)
- Federation with iPlant's authentication system:
- People will iPlant login credentials can log into CoGe as a registered user.
- Will enable the creation of personal data in CoGe
- Will enable more customization and saving of preferences for various tools in CoGe
- Will enable users to save particular analyses and datasets within CoGe
- Will enable import and export of data from CoGe to people's iPlant Data Store accounts
- Enhanced data security model:
- Will enable unpublished data to be restricted to a user or a group of users
Please come test the new CoGe: http://coge.iplantcollaborative.org and send Eric Lyons any problems you come across.
Since the holidays are coming and usage of CoGe tends to decrease, hopefully any bugs won't affect too many people while they are fixed. The migration of the domain names registered to CoGe will change once the server has been reasonably tested. Other CoGe services will migrate after that (e.g. this wiki).
CoGe domain names:
Pigeon-pea genome (Cajunus cajan) has been added to CoGe
Nov. 29th, 2011
The International Initiative for Pigeonpea Genomics has released the pigeon pea genome.
The pigeon-pea genomes may be accessed in CoGe: http://genomevolution.org/CoGe/OrganismView.pl?oid=34028
Please see this link for a syntenic dotplot between pigeonpea and medicago: http://genomevolution.org/r/49ua This syntenic dotplot has the syntenic gene pairs' evolution distance colored to differentiate orthologous and out-paralogous syntenic regions.
NCBI Genome Update: Over a thousand new genomes available in CoGe
Nov. 28th, 2011
The NCBI genome loading program for CoGe has been updated as is currently adding thousands of genomes from NCBI. Keeping CoGe current with all of the genomes at NCBI has been a challenge as their underlying data model for storing and organizing genomes evolves. The new program crawls all of NCBI's BioProjects searching for those with genomes and associated sequence. Prior to this data load there were approximately 12,100 genomes from 10,600 organisms. Approximately 40% of NCBI's BioProjects have been crawled and the current genome stats are:
Organisms: 12,093
Genomes: 13,969
Nucleotides: 305,480,720,992
Genomic Features: 99,814,749
Annotations: 224,582,292
For those that are curious, CoGe has maintained a MySQL DB transaction rate of 2000-3000 per second (majority writes/inserts) for the past 24 hours, thanks in no small part to its SSD configuration.
Note: After more performance monitoring, peak DB transactions top 9000 per second during heavy use from the genome loading programs and website activity.
Optical fun with CoGe
Nov. 22nd, 2011
Which direction does the DNA spin? Depending on how your mind is interpreting the dark and light colored dots of the DNA molecule as being "near" or "far", the helix can spin in both directions.
Thanks to Don McCarty for pointing this out.
Lamprey, Anole, and Frog genomes added/updated to CoGe
Nov. 19th, 2011
[www.ensembl.org Ensembl] version 64 genomes of Lamprey, Anole, and Frog have been added to CoGe:
Petromyzon marinus (lamprey): http://genomevolution.org/CoGe/OrganismView.pl?oid=30737 Xenopus (Silurana) tropicalis (western clawed frog): http://genomevolution.org/CoGe/OrganismView.pl?oid=33964 Anolis carolinensis (green anole): http://genomevolution.org/CoGe/OrganismView.pl?oid=33828
Both the unmasked and Masked versions of the genomes are available. For an example Syntenic dotplot between Xenopus and Tetraodon (pufferfish), please see: http://genomevolution.org/r/48w9
This dotplot uses the Syntenic path assembly to order and orient the contigs of Xenopus to the well assembled genome of Tetraodon (Frog versus Pufferfish): http://genomevolution.org/r/48w9
This dotplot uses the Syntenic path assembly to order and orient the contigs of Xenopus and Anolis (Frog V Green Lizard): http://genomevolution.org/r/48zk
Thanks to Bill Spollen for requesting these genomes.
Updated and New Plant Genome Resources
Nov. 10th 2011
The CoGePedia Sequenced plant genomes page has been updated with the latest published genomes, including the just published genomes of both pot and pidgeon pea! In addition, we have added two new pages that may be of interest to those who (like me) are constantly having to pull together introduction sections and can't remember what the right citation for well known genomic information is:
- Plant Genome Papers lists the papers describing every published plant genome, when and where it was published, and how much attention (in the form of citations) the various genomes have attracted so far.
- Plant paleopolyploidy is a list of known ancient whole genome duplications among the various plant species with sequenced genomes including information on when and how the whole genome duplications were discovered.
Both pages are clearly works in progress so please continue to contact us if we've missed genomes, whole genome duplications, or citations which should be on the list.
Main CoGe Database is down
Nov. 3rd 2011
7:00 (PCT USA) 14:00 (GMT)
Last night I ran a repair table on the main database for CoGe. This apparently ran into some problems and failed. I am currently hunting down the problem, and the main CoGe site is currently off-line. Technically, the tools are all available, but some of them are not working. The problem appears to be located in the "locations" table of the [CoGe database]. This table records the locations for all of CoGe's Genomic features. For anyone that needs to get some work done with CoGe, they are welcome to use the development server hosted at:
http://coge.iplantcollaborative.org
This version of CoGe has been under development to federate CoGe's user authentication system with the authentication system provided by the iPlant Collaborative. As such, there has been many code changes dealing with registered users and accessing restricted/private genomes. These changes are NOT fully tested and may cause some problems. Also, the development server is using an out-of-date version of the main CoGe database (though most of the genomes should be there). If you use the development server and run into any of these problems, please feel free to send Eric Lyons an email. I'd appreciate the reporting of any bugs as well as your patience with the current situation.
In case of catastrophic failure of the main database, please know that in addition to the development server, there is a full backup of the main CoGe database. These are generated weekly.
Also, thanks to Ben Field for notifying me of the problem. I deeply appreciate the help of community members in alerting me to problems with the site as well as suggestions for making it better.
Update: 8:00am
- Another "repair table" is being run on the main CoGe Database.
- Backup database is being restored on the dev server for CoGe (coge.iplantcollaborative.org). Once this is up and running, I'll point the main CoGe site to use this database and database server in case the main database has not yet been repaired.
Update: 9:30am
- backup coge database has been deployed to CoGe development server, currently undergoing "optimization" (want to avoid whatever happened to the main database)
Update: 5pm
- main coge database has been repaired. Warning and update messages taken down from the website. Let me know if anyone has any problems.
CoGe Tutorial Published in Maydica:
Oct. 24th 2011
A comprehensive open-access tutorial on using CoGe has been published in Maydica: http://www.maydica.org/articles/56_183.pdf
Abstract:
Of all the major plant groups, the grasses, with the complete genomes of five species, are the best positioned to take advantage of comparative genomics to obtain insight into functional genetic elements. Of all the grasses, maize is the best characterized in terms of genetics, development, and evolution. We provide several examples of how the web-based comparative genomics system CoGe may be used to aid in the interpretation of the maize genome sequence. These examples include verifying gene models, identifying differences between genome as- semblies, identifying conserved non-coding sequences, identifying syntenic regions between species and poly- ploidies, and identifying homeologs within maize and orthologs between maize and other grass genomes. In addition, a comprehensive list of orthologous gene sets is provided between maize and Sorghum, foxtail millet, rice, and Brachypodium.
While the article focuses on the maize genome as its primary genome, the methods are applicable to any genome.
Correction to the Classical Maize Gene and Syntelog List
Sept. 29th 2011
Phil Stinard identified an error in incorrectly assigning classical maize genes as being present in B73. Thanks to Mary Schaeffer for passing along this information and James Schnable for correcting these in the Classical Maize Gene and Syntelog List.
The following genes are now assigned as being not present in the B73:
- S
- lc1
- sn1
- hopi1
New options in SynMap
Sept. 12th, 2011
There are a couple of new options available in SynMap:
Force dotplot to be a square: You can find this option under the "Display Options" Tab with the line "Dotplot axes relations".
SVG Version of the Dotplot: There will be a new file, "SVG Version of the Syntenic Dotplot" to download in the "Links and Downloads" section of the results. This file will only appear if some form of synonymous rates are calculated and visualized (available under the "Analysis Options" tab").
Thanks to James Schnable for creating the SVG program for SynMap!
Potato genome added to CoGe
Sept. 3rd, 2011
Genome published: http://www.nature.com/nature/journal/v475/n7355/full/nature10158.html
The genome added was doubled the monoploid S. tuberosum Group Phureja clone DM1-3 516R44 (DM):
- unmasked: http://genomevolution.org/CoGe/OrganismView.pl?dsgid=12277
- masked: http://genomevolution.org/CoGe/OrganismView.pl?dsgid=12278
Please note: this version of the genome does not have annotations available.
Thanks to Will Spooner for the notification!
Brassica rapa genome added to CoGe
Sept. 3rd, 2011
Genome published: http://www.nature.com/ng/journal/vaop/ncurrent/full/ng.919.html#/group-1
Sequenced by: BGI
Brassica rapa has had a hexaploidy event subsequent to the most recent tetraploidy event in the Arabidopsis lineage.
Thanks to Will Spooner for the notification!
Cannabis sativa Pseudoassembled genome added to CoGe
Aug. 23rd, 2011
SynMap has the option to assembled one genome against another using syntenic. Such Syntneic path assemblies may be used to create a Pseudoassembly of a genome when only a contig level assembly exists. SynMap makes generating these Pseudoassemblies easy to do. Such a Pseudoassembly of the 175,000 Cannabis sativa genome was performed against the peach genome (read here to learn why peach was chosen). This pseudoassembly was reloaded back into CoGe and permits using CoGe's tools to compare the Cannabis genome at multiple levels of resolution.
To see this example: Cannabis sativa cultivar Chemdawg (marijuana)
Pseudoassemblies may be quite useful as more genomes are sequences on the cheap. Such sequencing project yield low-quality draft genomes that are usually assembled into several tens of thousands of contigs, and pseudoassemblies permit the rapid generation of large sequences that are easier to use in comparative genomic analyses.
Cannabis sativa cultivar Chemdawg (marijuana) added to CoGe
Aug. 22nd, 2011
The genome of the extremophile Cannabis sativa cultivar Chemdawg (marijuana) has been added to CoGe: http://genomevolution.org/CoGe/OrganismView.pl?oid=33804
This genome was sequenced by Medicinal Genomics (located in the Netherlands). It was sequenced with one lane of the Illumina HiSeq (2x100) platform and assembled with CLCbio’s workbench. Additional information about the assembly and genome may be found: http://www.medicinalgenomics.com/the-c-sativa-genome/
You can access Cannabis in CoGe: http://genomevolution.org/CoGe/OrganismView.pl?oid=33804
Cannabis is a member of the plant order Rosales. Of sequenced genomes in that order, the peach genome is a fantastic comparator. The reason for this is due to its high-quality sequence and assembly, and its genomic evolutionary history that does not contain any whole genome duplication event subsequent to the Eudicot paleohexaploidy shared by nearly all dicots (at least the eurosids and the astrids). As such, its genome structure is probably very similar to the common ancestor of order Rosales, and perhaps the eudicots as a whole. This likely ancestral state of the peach genome makes it quite suitable for generating a Pseudoassembly of highly fractured, low quality genome assemblies such as this Cannabis genome. CoGe's tool SynMap has an algorithm to tile contigs along any other "reference" genome in CoGe.
The Syntenic path assembly of Cannabis to the peach genome may be viewed: http://genomevolution.org/wiki/index.php/Syntenic_path_assembly#Cannabis_sativa_.28marijuana.29_v._Prunus_persica_.28peach.29
This shows the Cannabis genome sequence contains nearly the entire gene content of Peach.
Eutrema parvulum (Thellungiella parvula) added to CoGe
Aug. 17th, 2011
The genome of the extremophile crucifer Eutrema parvulum (Thellungiella parvula) has been added to CoGe: http://genomevolution.org/CoGe/OrganismView.pl?dsgid=12242
You can read about this genome in this Nature Genetics Letter: http://www.nature.com/ng/journal/vaop/ncurrent/full/ng.889.html
For a syntenic dotplot between it and Arabidopsis thaliana, please this SynMap anlaysis: http://genomevolution.org/r/3ws0
New Version of Setaria italica (foxtail millet) added to CoGe
Aug. 16th, 2011
Version 2.1 of Setaria italica has been added to CoGe. This genome was obtained from JGI/phytozome: http://phytozome.net
Unmasked version: http://genomevolution.org/CoGe/OrganismView.pl?dsgid=12240 Masked version: http://genomevolution.org/CoGe/OrganismView.pl?dsgid=12241
Thanks to Gina Turco for the request.
New Version of Fragaria vesca (woodland strawberry) added to CoGe. This time with gene models!
Aug. 11th, 2011
Version 1.1 of Fragaria vesca (woodland strawberry) has been added to CoGe http://genomevolution.org/CoGe/OrganismView.pl?dsgid=12186 .
This version contains gene models with permits more fun with syntenic dotplots: http://genomevolution.org/r/3wdb
This dotplot is strawberry versus peach. Besides from be a great summer fruit salad, this dotplot colors syntenic gene pairs based on their synonymous mutation values. From it, it is easy to see neither genome has had an independent whole genome duplication since the Eudicot paleohexaploidy event.
Thanks to Aaron Liston for requesting this genome.
Daphnia pulex (common water flea) added
Aug. 3rd, 2011
You can get all your water flea genomics here: http://genomevolution.org/CoGe/OrganismView.pl?oid=33760
Thanks to Mike Freeling for the request.
Several bugs fixed as a result of the code update
July 29th, 2011
Additional bugs were squashed today due to the major code update to CoGe's internal services. Part of the update included further modularization of the web-services from backend services. A few programs the ancillary support programs for CoGe's web-services were not correctly being passed the base configuration file for a given web-deployment and were therefore crashing. This has been corrected, but please email Eric Lyons if any problems are encountered.
Update to GenomeList
July 29th, 2011
GenomeList has been updated to:
- include a link back to GenomeList for selected genomes. This is useful if a broad selection of genomes was made and needs to be refined.
- include a link to easily download a fasta file for a given genome
- include a link to coge_gff to generate a gff file of all Genomic features and annotation in a genome
- include a TinyURL link to regenerate the genome list. This link is found at the top of the genome list.
Example GenomeList link: http://genomevolution.org/r/3v8n
Major code update to CoGe
July 27th, 2011
CoGe has undergone a major update of its web-based system today. A few bug fixes and feature enhancements mixed in, with the major one being the addition of GenomeList for creating a list of genomes, getting an overview of their genomic content, and then sending the list to other tools (e.g. CoGeBlast).
Behind the scenes was a further modularization of the web-interface from the backend support services and modules. The primary reason for this is to enable to creation of multiple CoGe installations. There has been a few requests by people for a clade/group of organisms specific installation of CoGe. With iPlant's cyberinfrastructure support, this should be possible (providing the code-base supports it).
There were some sticking points this morning migrating server specific changes from the iPlant development server to the main CoGe server, but hopefully this didn't affect too many people. However, there is a high-likelihood of additional bugs in the system that I failed to catch! Please email Eric Lyons if you find any problem.
Otherwise, we are hoping to make a full migration to iPlant's resources in the near future. iPlant's coge server is being upgraded with some additional attached storage for continual growth of the platform.
Weill's Date Palm genome version 3 has been added to CoGe
July 4th, 2011
You can find its genome in OrganismView: http://genomevolution.org/CoGe/OrganismView.pl?dsgid=11942
And a Syntenic path assembly to rice here: http://genomevolution.org/r/3ox8
This is a very rough genome (50,000+ contigs; the largest is 470KB; 13 larger than 300KB). However, the syntenic path assembly in SynMap with the option to remove any contig that doesn't have a syntenic signal makes identifying sytnenic regions a breeze (see the above link).
See this example of micro-synteny as seen in GEvo: http://genomevolution.org/r/3oxa
Thanks to: Haibao Tang, Devin O'Connor, and Jim Leebens-Mack for requesting this genome.
July 14th, 2011
The masked version of the Palm genome has been created and added to CoGe: http://genomevolution.org/CoGe/OrganismView.pl?dsgid=11947
Thanks to Haibao Tang for providing the masking procedure.
JGI's Eucalyptus grandis BRASUZ1 has been added to CoGe
June 30th, 2011
You can find its genome here: http://genomevolution.org/CoGe/OrganismView.pl?oid=33537 (masked and unmasked sequence)
With a comparison to the peach genome, Eucalyptus looks to have had its own whole genome duplication subsequent to the Eudicot paleohexaploidy: http://genomevolution.org/r/3ol1
Thanks to Josquin Tibbits for recommending this genome!
Arabidopsis thaliana resequenced genomes (C24, Bur-0, Kro-0, Ler-1) from 1001genomes.org has been added to CoGe
June 30th, 2011
The "High Quality" sequences generated by the 1001genomes project for the resequencing of several arabidopsis strains has been added to CoGe. This includes:
While these genomes contain many contigs, CoGe's Syntenic path assembly algorithm can arrange and orient them against the reference genome Col-0: http://genomevolution.org/r/3okf
Thanks to Maggie Woodhouse for this suggestion!
OrganismView's Feature List display updated
June 22nd, 2011
OrganismView has a minor update for where the lists of Genomic features are displayed. The old version would display the summary list of genomic features below all the information panels. This would mean that each time a summary list was generated, it would replace the prior one. For example, if you retrieved the list first for the entire genome and second for a particular chromosome. Now, each information panel's genomic feature list appears to the right of the information summary. This allows the entire genome's feature list to be display simultaneously with the chromosome's feature list.
Broad Institute's Coccidioides group Database added to CoGe
June 21st, 2011
The entire set of sequences and associated annotations for Coccidioides has been added to CoGe. These soil fungi are pathogenic and can cause coccidioidomycosis, aka valley fever, in humans. The original data may be obtained from: http://www.broadinstitute.org/annotation/genome/coccidioides_group/MultiHome.html
And accessed through OrganismView: http://genomevolution.org/CoGe/OrganismView.pl?org_desc=Coccidioides
Thanks to Marc Orbach for suggesting and locating these genomes.
UC Berkeley Data Center Back Online
June 12th, 2011
The UC Berkeley Data Center power upgrade went smoothly. CoGe has booted up and is back online.
Thanks to:
- James Schnable for being on duty to bring CoGe down and back up.
- The entire team at the UC Berkeley Data Center for completing such a complicated upgrade to their Center and for continually updating their clients as to the progress of the operation.
CoGe Downtime June 12th, 2011
June 3rd, 2011
CoGe will be down on June 12th due to maintenance on the power infrastructure at the UC Berkeley Data Center. We will do our best to bring CoGe back up as soon as possible.
Here is their announcement:
Description: The [UC Berkeley] campus data center has been a valuable resource for campus computing for the past seven years. Demand for this highly secure, highly available, and network-redundant facility continues to rise. The current facility has reached its power and cooling capacity and Capital Projects has initiated a major renovation project intended to increase each of these capacities, while also integrating newer, more efficient systems to help the campus achieve its long-term energy conservation goals.
As part of this effort, the replacement of some core components of the data center’s power infrastructure is required. For safety reasons, a full power outage to the data center is scheduled for Sunday, June 12, 2011, from 7:00 am to 3:00 pm. The data center will rely entirely on outside air, rather than air conditioning, to provide cooling for the duration of this period. A minimal number of systems with broad campus impact, including CalMail, CalAgenda, and the campus home page, will be provided with temporary power during this outage. In the unlikely event that the data center air temperature exceeds a level appropriate for the safe operation of equipment, some of these systems may need to be shut down as well.
The list of widely used systems that are intended to remain available is below. This list is still being finalized, so additional systems may be added as campus needs require. This list will not include systems for which departments have made separate arrangements.
Citrus genomes added
May 6th 2011
The genomes of:
- Citrus clementina (Clementine mandarin): http://genomevolution.org/CoGe/OrganismView.pl?oid=33274
- Citrus sinensis (citrus, Sweet orange): http://genomevolution.org/CoGe/OrganismView.pl?oid=33273
Have been added to CoGe. These were sequenced by JGI.
A quick syntenic analysis of sinensis to peach shows that it appears to have no subsequent whole genome duplication event to the eurosid Paleohexaploidy: http://genomevolution.org/r/2zdv
Sequenced Plant Genome Phylogeny Update
May 6th 2011
James Schnable has updated the phylogeny of angiosperms for sequenced plant genomes.
CoGe Workshop at Berkeley
Apr. 19th 2011
Here is the outline/syllabus of the workshop help at Berkeley hosted by the iPlant Collaborative, the Department of Plant and Microbial Biology, QB3-CGRL (Computational Genomics Resource Laboratory), ARS-Plant Gene Expression Center, and the Freeling lab: 2011 Berkeley Workshop
This outline contains links to specific analyses used in the workshop.
Horizontal Genome Transfer
Mar. 31st 2011
Here is a fun example of a mitochondria genome being inserted into a plant chromosome: Horizontal transfer of mitochondria genome: Horizontal transfer of mitochondria genome
Second "Run GEvo Analysis!" button added to GEvo
Mar. 29th 2011
For those times when scrolling to the top of the screen to find the "Run GEvo Analysis!" button is too much work, a second button has been added at the bottom of the configuration box. This is quite useful when comparing >6 genomic regions.
Thanks to David Braun for this suggestion!
Bug Fix in FeatView
Mar. 29th 2011
Thanks to Damon Lisch for pointing out a bug in FeatView that was exposed by Firefox v4. This bug was also affecting Google Chrome (but not Safari). Please let Eric Lyons know of any problems you have running Firefox v4 (or other problems in general).
New tutorial for performing genomic rearrangement analyses
Mar. 11th 2011
A new tutorial has been written for showing how to figure SynMap to generate a link to GRIMM (by Glenn Tesler, University of California, San Diego) for performing genomic rearrangement analysis.
Tutorial: How to perform a genomic rearrangement analysis
SynMap now has support for BlastP
Mar. 7th 2011
You can now select to compare protein sequences between genomes with annotated protein coding features (CDS).
Thanks to Angelique D'Hont for the suggestion.
Cochliobolus heterostrophus C5 from JGI loaded into CoGe
Mar. 2nd 2011
You can find Cochliobolus heterostrophus C5 in OrganismView: http://genomevolution.org/CoGe/OrganismView.pl?dsgid=11258
Both masked (by JGI) and unmasked version of the genome are available.
For a syntenic dotplot between C. heterostrophus to Pyrenophora tritici-repentis strain Pt-1C-BFP (the closest relative I could find in CoGe) please follow: http://genomevolution.org/r/2m0n
This is a neat syntenic dotplot showing extensive synteny and intrachromosomeal rearrangements (though these are both contig level assemblies).
Thanks to Daniel Lawrence for request.
Sort chromosomes by name in SynMap
Feb. 26th 2011
After a couple of requests, SynMap now has an option to sort chromosomes by name instead of by size. You can read how to set this option here.
Thanks to:
- Angélique D'Hont from CIRAD
- James Schnable from UC Berkeley
for this suggestion.
How to load genomes into CoGe
Feb. 22nd 2011
If you have a CoGe installation, access to the main CoGe server, or just curious to know what is needed to load a genome into CoGe, here is a page on How to load genomes into CoGe. This is all run from the command line, and when CoGe's user permission data management system matures, this procedure will be made available via the web.
Giant Panda genome loaded into CoGe
Feb. 19th 2011
You can see the genome in CoGe at: http://genomevolution.org/CoGe/OrganismView.pl?dsgid=11227
This was one of the first big genomes sequenced using only Next Generation Sequencing Technology and assembled De novo. As a result, the assembly is rather poor compared to a fully assembled genome like the dog genome. However, through comparative genomics with SynMap, identifying syntenic regions and determining that nearly full coverage was obtained is as easy as a few mouse clicks: syntenic path assembly of the WGS panda genome to the fully sequenced dog genome. This will be quite useful as more and more large genomes are sequenced using these techniques (fast, cheap, and still very useful!)
First Metagenome loaded into CoGe
Feb. 19th 2011
Technically, there is no reason why CoGe can't store metagenomes. Its core data model stores a collection of sequences that, thus far, has been organized into a genome, but can accommodate any collection of sequences. So the first metagenome was loaded into CoGe from NCBI:
Mine drainage metagenome, whole genome shotgun sequence
And can be seen in CoGe: http://genomevolution.org/CoGe/OrganismView.pl?oid=32988
Assembling contig-level assembles to a reference genome using synteny
Feb. 18th 2011
SynMap has an option for generating a Syntenic path assembly with the click of a button. When complete, there is an option to print out your assembled genome.
CoGe 2011 Plant and Animal Genome conference presentations available for download
Feb. 10th 2011
For a complete list of PAG sessions: http://www.intl-pag.org/19/19-workshops.html
"CoGe: Comparative genomics made easy!"
Eric Lyons, iPlant Collaborative and University of Arizona, Tuscon AZ (ericlyons@e-mail.arizona.edu)
PDF available at: http://genomevolution.org/CoGe/data/distrib/presentations/PAG-2011-CoGe-CompG.key.pdf
"10,000 Genomes at Your Fingertips"
Eric Lyons, iPlant Collaborative and the University of Arizona, Tuscon AZ (ericlyons@email.arizona.edu)
PDF available at: http://genomevolution.org/CoGe/data/distrib/presentations/PAG-2011-CoGe-ComputerDemp.key.pdf
Chocolate genome gene models added
Feb. 4th 2011
Thanks to CIRAD for sharing their cacao gene models. These have been added to the Theobrama cacao genome in CoGe: http://genomevolution.org/CoGe/OrganismView.pl?dsgid=10997 .
For an example of how these gene models may be used in whole genome comparisons, see this analysis between chocolate and peach: Chocolate-peach syntenic dotplots. It shows how the evolutionary distance between sytnenic gene pairs may be visualized to differentiate between Orthologous syntenic regions derived from the divergence of these lineages, and Out paralogous syntenic regions derived from their shared Paleohexaploidy ancestry.
Arabidopsis thaliana TAIR version 10 has been added!
Jan. 27th 2011
Version 10 of the Arabidopsis thaliana genome has been added to CoGe: http://genomevolution.org/CoGe/OrganismView.pl?dsgid=11022
Thanks to all the work by the folks at TAIR
For a syntenic dotplot of version 9 versus version 10 of Arabidopsis thaliana (with the evolutionary distances of syntenic gene pairs calculated) see: http://genomevolution.org/r/2hiz
Chocolate genome added: from the International Cacao Genome Sequencing Consortium
Jan. 26th 2011
The genome of Theobroma cacao has been published: http://www.nature.com/ng/journal/vaop/ncurrent/full/ng.736.html
You can view this genome in CoGe at: http://genomevolution.org/CoGe/OrganismView.pl?dsgid=10997
To view some Syntenic dotplots of Cacao: Cacao syntenic dotplots
Of note, this genome has not had any whole genome duplication events since the Paleohexaploidy event at the base of the eurosids.
Version 2 of the Maize Genome, Now With Gene Models
28 November 2010
Both the 50x super masked and unmasked versions of the B73_refgen2 maize genome are now updated with the new gene models released by maizesequence.org over thanksgiving break. The new genome annotation consists of 110,028 genes, many with alternative transcripts, which can be broken down as followes:
- 29,082 transposon related genes
- 17,615 putative pseudogenes
- 63,276 "real" genes. Please note while these genes were annotated as "protein coding" in the current release, they include predicted microRNA genes.
Maintenance Complete
Sept. 16th 2010
CoGe's servers have successfully be moved to a new rack space. Thanks to James, Bao, and Brent for making this happen.
Pending CoGe Maintenance
Sept. 15th 2010
We have received word from the UC Data Center which houses CoGe that we need to move our servers to a new rack space. This should only take an hour or two. Our tentative schedule time for the move is:
Sept 16th 2010 at 1pm (PCT)
We apologize for any inconvenience this may cause any of CoGe's users.
SynMap update
Aug. 26th 2010
Organisms selected in SynMap have links in their taxonomic descriptions. If you click on a term in the taxonomic description, that term is automatically entered into the organism description search. All organisms with a matching taxonomic term will be displayed. This makes it faster to find organisms related to the one in which you are interested.
OrganismView update
Aug. 26th 2010
OrganismView now has more links for finding information about an organism, and to internal CoGe tools.
External searches under organism information:
- NCBI
- Wikipedia
Internal CoGe links:
- CodeOn: automatically generates a table of amino acid usage as a function of the GC content of CDS sequences.
- SynMap: (under Genome information) automatically loads SynMap with both genomes specified to the one selected. This makes is quick to start generating whole genome comparisons and Syntenic dotplots.
Home Page update
Aug. 26th 2010
CoGe's homepage menu "Latest Genomes" now has links to search for the organism name in
- CoGe's OrganismView
- NCBI
- Wikipedia
This makes it quicker to find information on an organism, specifically if you have no idea what it is. Helpful considering that there are nearly 9,000 organisms in CoGe.
CoGeBlast update
Aug. 26th 2010
CoGeBlast now has support for specifying blastn, tblastx, lastz, megablast, and discontinuous megablast when searching with nucleotide sequences.
10,000th genome loaded!
Aug. 4th 2010
Brassica rapa has been added to CoGe and represents the 10,000th genome loaded in CoGe. Its sequence was generated by the BGI located in China. This relative of Arabidopsis is a wonderful addition to sequenced plant genomes. Their lineage share a series of whole genome duplication events (commonly known as alpha, beta, and gamma -- the latter happening prior to the radiation of the eudicots). Since their divergence, Brassica rapa has had a triploidy while Arabidopsis has had none.
Genome update from NCBI
June 28th 2010
A new update of genomes from NCBI has finished. This includes genomes from all domains of life. CoGe now has genomic sequence from 8,872 organisms comprising 9,999 genomes. There is also a new option on the homepage to list the most recently added genomes.
SIP 2010 workshop syllabus
June 23rd 2010
The syllabus for a day-long workshop on how to use CoGe for the Society for Invertebrate Pathology's conference (SIP 2010) is now available. This workshop focuses on:
- Getting an overview of how CoGe is designed for allowing scientists to create their own open-ended analyses
- Learning what the various tools in CoGe do and they to use them
- Working through specific sets of example problems focused on analyzing two groups of organisms important for invertebrate pathology: baculoviruses and Bacillus thuringiensis
The workshop's syllabus is available: SIP2010
CoGe's update progress
June 18th 2010
The switch to the new server went as smoothly as I could have hoped.
Besides from new hardware (which should greatly accelerate many of CoGe's analyses and improve system stability), this installation welcomes a new version of CoGe too!
This new version of CoGe has:
- Update UI
- Various feature extensions on existing tools
- Updated algorithms (new blast API with support for the megablast families, LastZ)
- New database additions
- Update of core modules for database API
- New configuration files that will help deployment of CoGe to new sites
Please contact Eric Lyons if you find any bugs!
Today is the day
June 17th 2010
Going to through the switch today. Expect some downtime with CoGe and some support systems being temporarily off line.
New CoGe Server Update
June 10th 2010
It appears that most of the software updates and migration to the new server are working. We have deployed the new server to the UC Data Center, but due to some complications with rack-space, IP address allocation, sub-nets, firewalls, etc., things may be in flux for a while. We've had to take our development server (aka toxic) off line and put the new server on its IP address till those things get sorted out. In the meanwhile, we will plan on making the switch to production on the new server soon (hopefully next week). When this happens, expect CoGe to be offline for a couple of hours, but we will do our best to keep downtime to a minimum.
New CoGe Server is being readied!
June 2nd 2010
We have our new server for CoGe! Its deployment will not only include new performance improvements due to more computing power, but all several changes and additions to CoGe:
- new user interface
- new algorithm options
- new structure of the underlying code-base to make it is easier to redeploy (in anticipation of eventually getting the code-base released to those interested)
We are planning on moving the new server to the UC data center this Fri. After some more testing and bug hunting, we will switch our current production server's IP address to this machine. There is a high chance that there will be some downtime for CoGe during this switch and we will post announcements as to when this change will happen! In the meanwhile, if anyone is interested in testing new CoGe, please e-mail Eric Lyons.
SGRP: (Sanger Institute) yeast genomes added to CoGe
May 18th 2010
75 Yeast genomes from SGRP (Saccharomyces Genome Resequencing Project) have been added to CoGe. For a complete list of Organisms, please see SGRP: Sanger Institute Yeast Genomes.
CoGe post on The OpenHelix
May 5th 2010
Eric Lyons wrote a piece about CoGe for The OpenHelix Blog
Version 2 of Maize B73 genome added to CoGe
May 3rd 2010
This release does not yet have annotations (yet)!
You can view the genome in CoGe at: http://genomevolution.org/CoGe/OrganismView.pl?dsgid=9106
This sequences was obtained from: http://www2.genome.arizona.edu/genomes/maize
And can read about differences in the assembly between versions 1 and 2: here.
Version 2 of Vitis vinifera (grapevine) genome added to CoGe
Apr. 10th 2010
You can view the genome in CoGe at: http://genomevolution.org/CoGe/OrganismView.pl?dsgid=9048
Version 2 with 12x coverage was obtained from Genoscope.
There are some changes to the assembly with new contig orders and additional sequence added to the pseudomolecules which can been seen here.
New NCBI Genome Update. CoGe surpasses 8,900 genomes from 8,200 organisms
Apr. 9th 2010
Finished an update from NCBI. However, this is not a complete listing of all genomes available at NCBI due to some API problems getting some genomes. You can read about this problem below.
Version 3 of Medicago truncatula added to CoGe
Apr. 9th 2010
You can view the genome in CoGe at: http://genomevolution.org/CoGe/OrganismView.pl?dsgid=8976
Syntenic comparison of version 3 to version 2 shows extensive changes in the primary sequence. Some chromosomes have had their sequence substantially updated.
Prunus persica (peach tree) added to CoGe
Apr. 9th 2010
You can view its genome in CoGe at: http://www.genomevolution.org/CoGe/OrganismView.pl?oid=30980
Its genome was produced by the International Peach Genome Initiative and its sequence was obtained from phytozome. This genome is currently unpublished and therefore under the publication restrictions of the Fort Lauderdale Convention.
Peach is a eudicot in the Rosaceae family.
Automatic NCBI Genome Loader Update
Apr. 8th 2010
The automatic NCBI genome loader is running today. It has been a while since I last ran it after running into an API problem with NCBI's eutils tools three months ago. The issue is still unresolved and even after checking in every two weeks for a status update, I have yet to receive any word as to when the bug will be fixed. For those interested, here is my bug report sent at the end of January:
Issue (http://jira.be-md.ncbi.nlm.nih.gov/browse/HD-1843): Key: HD-1843 Summary: Unable to get some genomes using eutils Type: Task Status: In Progress Priority: Normal Assignee: Matten, Wayne Reporter: Nobody Description: Hi, I've be checking which genomes are available from NCBI using eutils by getting a list of all the genome project ids (genomeprj) and then retrieving their associated genome ids. I've found that a lot of the recently deposited genomes (usually with accessions CPXXXXXX) are have a genomeprj id but no associated genome id. For example, genomeprj=30031. It is listed in this list: http://eutils.ncbi.nlm.nih.gov/entrez/eutils/esearch.fcgi?db=genomeprj&term=all%5Bfilter%5D&retmax=999999 But has no genome id: http://eutils.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?db=genome&dbfrom=genomeprj&id=30031 However, it does have an entry in genbank: http://www.ncbi.nlm.nih.gov/nuccore/CP001637.1?ordinalpos=3&itool=EntrezSystem2.PEntrez.Sequence.Sequence_ResultsPanel.Sequence_RVDocSum I am probably missing something obvious. Can you help me figure out how to get a list of all the genomes at NCBI? I am using these data in an NSF funded and publicly available comparative genomics platform (http://synteny.cnr.berkeley.edu/), and have programs that check for new genomes and new versions of existing genomes from NCBI on a periodic basis. It is important for this system to be as up to date as possible with regards to the large number of genomes that are becoming available as there are many researchers using this tool for their work. Thanks in advance for your help, -Eric Lyons
If anyone has any solutions to this problem, please contact me.
Major bug fix in SynMap
Mar. 26th 2010
While testing the prior bug fix, I discovered that SynMap wasn't working on genomic sequence comparisons (as opposed to CDS sequence comparisons). This was due to the new analytical pipeline's data processing requiring unique names for each blast hit. Otherwise, multiple hits to the same sequence name would get removed as a Local duplicate. As all hits to a genomic sequence were named according to the chromosome, all such hits were flagged as Local duplicates and removed from the analysis.
As always, if you find a problem in CoGe, feel free to email Eric Lyons and let him know what you've found. There are now too many options and buttons to click in CoGe for me to test with each update.
Minor bug fix in SynMap
Mar. 25th 2010
With SynMap's new analytical pipeline, there are still some bugs to be worked through. Hopefully got one today in the script that converted blast input files to bed format, which is required for the program to find local duplicates in the compared genomes. These local duplicates are removed from the algorithm for finding collinear series of putative homologous genes used to infer syntenic regions. Also, these local duplicate files are displayed in the download section of the results in case they are wanted for other analyses.
Hosting local tiny URL encoding
Mar. 24th 2010
Replaced using tinyurl.com for a local installation of a URL hashing and redirecting service. Makes generating these faster and allows for customized names. Note: the tinyurls will still work.
Sequenced plant genomes
Mar. 13th 2010
James Schnable has created a page detailing all of the sequenced plant genomes including:
- overview of their genomic content
- publications
- status of completion
- interesting factoids (e.g. The average US American eats 25lbs of bananas a year.)
Read about them here: Sequenced plant genomes
The JGI's Manihot esculenta (cassava) genome has been added
Mar. 13th 2010
This genome from the JGI brings CoGe up-to-date with phytozome v5.0.
You can access cassava in CoGe here, and get more information from phytozome.
The JGI's Cucumis sativus (cucumber) genome has been added
Mar. 12th 2010
You can access it in CoGe here. Or get more information about it from phytozome. This is apparently a distinct sequence from the one in Nature Genetics last November. That sequence was from "'Chinese long' inbred line 9930" this version comes from the inbred Gy14. More details here
SynMap updated
Mar. 12th 2010
After a month of work, SynMap has undergone several significant changes, incorporating new algorithms written by Haibao Tang and Brent Pedersen:
- new merging function for overlapping and neighboring diagonals (program: quota alignment)
- new method for detected tandem gene duplicates
- better reporting of all intermediate files used in the analysis, including tandem duplicates
These changes have also hoped to increase the stability of SynMap, which due to its long pipeline, has been known to crash for some genomes and/or specific parameter configurations. Please let Eric Lyons know if you have any problems with an analysis. Please send along the names of the organisms/genomes compared and a copy of the log file produced by each SynMap run (if possible).
Persistent GEvo bug fixed
Mar. 11th 2010
A long-stranding, but intermittent and annoying bug in GEvo has finally fixed. This (hopefully) solves the problem where once in a while, GEvo will return blank results to its interactive viewer, Gobe. The crux of the bug, and why it was intermittent (and hence difficult to reproduce and trouble-shoot), was a race condition between asynchronous client javascript code and server perl code. Perl was responsible for generating a random session id for the analysis, but it occasionally failed to return that id to the client code before the analysis was sent back to the server for processing. When this happened, the processing analysis received a default id and multiple analyses could be merged if the default id had been used within that past 24 hours (the length of time an analysis stays on the server before being deleted). When Gobe tried to process the results, the stored data and what was specified for initialization did not match, thus causing gobe to fail and return blank results. The solution: have javascript generate the analysis session id so there is no chance of a delay before the analysis is sent to the server for processing.
However, if anyone does come across this bug again (or any others), please let me know: Eric Lyons
Rice Version 6.1 loaded
Mar. 10th 2010
You can view it in GenomeView. This was retrieved from MSU's Rice Genome Annotation Project.
The classic set of Maize Genes
Mar. 9th 2010
James Schnable manually evaluate ~460 classic maize genes available from MaizeGDB and NCBI, determined their genomic positions in the maize genome, and found their Syntenic regions within maize (from its most recent Whole genome duplication event), sorghum, rice, and brachypodium. This list contains links to compare these syntenic regions using GEvo.
New plant genomes in CoGe
Feb. 10th 2010
Mimulus guttatus (monkey flower): http://synteny.cnr.berkeley.edu/CoGe/OrganismView.pl?oid=30760 Mimulus is an outgroup to the rosids (in the sister group, the asterids)
Populus trichocarpa (Poplar; cotton wood): http://synteny.cnr.berkeley.edu/CoGe/OrganismView.pl?oid=324 Version 2 of poplar!
Both are from the JGI.
MaizeGDB links to GenomeView
Feb. 8th 2010
MaizeGDB is now linking to CoGe's GenomeView so maize researchers can find maize-sorghum Syntenic gene sets and quickly perform syntenic analyses using GEvo. For an example view from MaizeGDB's genome broswer:
http://gbrowse.maizegdb.org/cgi-bin/gbrowse/maize/?name=chr1:1000000..1200000
For instructions on how to perform this workflow: MaizeGDB and CoGe
For more information on maize-sorghum syntenic analyses: Maize-Sorghum genome analyses
For a quick video walk through of the new connections: MaizeGDB_and_CoGe.27s_Maize-Sorghum_Orthologies
Syntelog visualization in GenomeView
Feb. 5th 2010
GenomeView has been updated to auto-detect Genomic features with annotations that are links to GEvo. These links provide an analysis of a Genomic feature (e.g. gene) to previously identified Syntologous sets of features. Currently, this has been implemented using syntelogs from maize and sorghum, but with the code in place, we will expand annotations for genomic features from other organisms for which we generated syntologous gene sets. For an example of this visualization in GenomeView please see: | this GenomeView of sorghum. Also, for an expanded list of glyphs used in GenomeView please refer to these examples.
Easy exporting and downloading of genomes
Jan. 16th 2010
OrganismView has new options for easily downloading the sequences of a genome in fasta format and retrieving all of its annotations in an GFF file. To access, just search for an organism and genome of interest, and look for the links under "Genome Information".
FastaView is linked to phylogeny.fr for one-click phylogenetics
Jan. 10th. 2010
We've linked to phylogeny.fr for quick and easy phylogenentic tree reconstruction. Now, you can build a list of fasta sequences and display them in FastaView, select protein or DNA sequences, edit them if necessary (e.g. add or remove sequences manually), and press a button to send them off to phylogeny.fr for:
- multiple sequence alignment (MUSCLE)
- maximum likelihood phylogenetic tree reconstruction (PhyML)
- tree visualization (TreeDyn)
For an example, use this link to FastaView and press the button "phylogeny.fr" at the bottom of the screen.
Special thanks to Haibao Tang for pointing out this incredible web resource!
Haibao Tang joins the Freeling lab
Jan. 4th 2010
Haibao Tang, an expert in plant comparative genomics and genome evolution, as well as a great python programmer, has joined the Freeling lab. His input and contributions will be most valued!
New Tutorials added
Jan. 4th 2010
New Tutorials have been added:
- How to find syntenic regions between genomes
- How to find inversions
- How to find rarely and frequently used codons in a genome
- How to generate an amino acid usage table for a genome
- Using synonymous mutation rates in SynMap to rapidly identify different whole genome evolutionary events
- How to extract all gene sequences from a genomic region
- How to identify putative horizontal gene transfer events
Linked to ProSite for protein domain searching
Dec. 24th 2009
FastaView is now linked to ProSite when viewing a protein sequence for protein domain searching. See this FastaView example and click on the link at the bottom of the page.
Improved implementation of DAGChainer in SynMap
Dec. 15th 2009
Thanks again to Brent Pedersen for some fantastic programming. He discovered that DAGChainer's C++ code's makefile did not include the -O3 optimization, rewrote the input/output methods of the compiled binary to read from STDIN instead of a file, and rewrote the perl front-end in python. Together, these changes increase CoGe's DAGChainer implementation in SynMap between 2-4 fold.
You can download his code at: svn co http://bpbio.googlecode.com/svn/trunk/scripts/dagchainer
CoGe Workshop being taught at SIP 2010
Nov. 30th 2009
Genomics: What every invertebrate pathologist needs to know. http://www.sip2010.org/index.php/Bioinformatics-Workshop.html
CoGe on OpenHelix and James and the Giant Corn
Nov. 18th 2009
Phillipe Lamesch from TAIR passed along a link to openhelix.com highlighting CoGe's tool GEvo. They put together a nice video showing GEvo. They, in turn, found this on a posting at the blog of James and the Giant Corn who had used GEvo for a grant proposal.
Maize Pseudomolecule Assembly with Gene Models Released
Oct. 20th 2009
Thanks to maizesequence.org for providing the sequence and annotations. The current pseudomolecule assembly of maize has been loaded into CoGe.
- Link to OrganismView for complete set of gene models.
- Link to OrganismView for filtered gene model set.
- Maize-Sorghum syntenic dotplot with syntologs colored by synonymous rate change.
- Maize-Maize syntenic dotplot with syntologs colored by synoonymous rate change.
CoGe surpasses 7000 organisms in its database!
Sep. 25, 2009
More fun for everyone!
NCBI Genome Loader Updated
Sep. 23, 2009
CoGe's automated NCBI genome loader has been updated and is once again checking NCBI regularly for new and updated genomes. You can get a snapshot of the number or organisms and genomic sequence in CoGe by checking its homepage, search for your genome of interest using OrganismView.
CoGe is linked to TARGeT: Tree Analysis of Related Genes and Transposons
Aug. 26, 2009
You can send a set of fasta sequence generated by FastaView directly to TARGeT.
New version of Gobe release!
July 21, 2009
Read general announcement Gobe. Major feature: transparent wedges are drawn to connect regions of sequence similarity.
Version 3 of CoGe is released!
July 15th, 2009
Read general announcement CoGe version 3.