Difference between revisions of "Flanking Gene Method"
(→Heading) |
|||
| Line 1: | Line 1: | ||
| − | + | [[Image:Flanking gene method.png|thumb|600px|right|[[GEvo]] analysis showing the flanking gene method.]] | |
| − | + | The flanking gene method is a procedure which compares two neighboring [[syntenic]] gene pairs in order to identify transposed or deleted genes. An example visualization of this method is shown to the right. Here, there are two pairs of syntenic genes (numbered 1 and 2). | |
| − | + | ||
| − | + | ||
| − | + | Kieran, go wild. | |
| + | |||
| + | |||
| + | the location of two sequential genes in a region orthologous between two species such that, given a pair of genes 1 and 2, if gene X is present between the two genes in one species but not the other (such that Species A=1 X 2, Species B=1 2), gene X is denoted as a possible transposed gene (as previously described in Freeling et al 2008). An outgroup (Species C) is used to distinguish between gene transposition in Species A and gene deletion in Species B. | ||
Revision as of 09:35, 20 April 2010
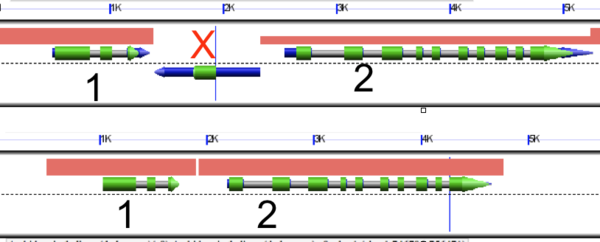
The flanking gene method is a procedure which compares two neighboring syntenic gene pairs in order to identify transposed or deleted genes. An example visualization of this method is shown to the right. Here, there are two pairs of syntenic genes (numbered 1 and 2).
Kieran, go wild.
the location of two sequential genes in a region orthologous between two species such that, given a pair of genes 1 and 2, if gene X is present between the two genes in one species but not the other (such that Species A=1 X 2, Species B=1 2), gene X is denoted as a possible transposed gene (as previously described in Freeling et al 2008). An outgroup (Species C) is used to distinguish between gene transposition in Species A and gene deletion in Species B.