CoGepedia:Current events: Difference between revisions
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
== | == CoGeBlast Update == | ||
Oct. 15th, 2012 | |||
[ | The CoGeBlast user interface was revamped for a cleaner appearance and simpler use. The functionality remains unchanged, except the addition of a button to import target genomes from existing [http://genomevolution.org/CoGe/Lists.pl lists]. | ||
<br> | |||
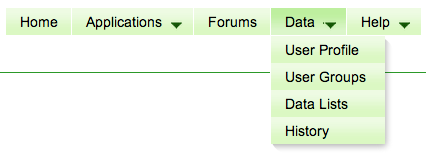
== CoGe "Data Tab" == | |||
Oct. 5th, 2012 | |||
With the roll-out of CoGe v5 comes the ability for users to more easily organize and share their data of interest. Most of these features are found in the "Data" tab in the CoGe menu located in the upper right part of the screen: | |||
*9am: We shut down the website at 9am and started the final backup and freezing of existing data and analyses. | [[Image:Screen shot 2012-10-05 at 9.19.34 AM.png]] | ||
*10am: Database is being replicated to the iPlant Data Store and copied to a backup server for processing and conversion to new database scheme. | |||
*11am: All web-code and libraries were backed up and new code deployed | *User Profile: Shows what information CoGe's stores about you (user name, real name, email address) and a list of your groups | ||
*12pm: updating database | *User Groups: Groups of users to which you have access. These groups are used to share lists of data | ||
*1pm: copying database back to iRODS | *Data Lists: Lists of data (genomes, features, experiments) to which you have access. May be private or public data. It is through these lists and User Groups that allows you to share private data with collaborators | ||
*2pm: copying database to coge server | *History: CoGe has always generated tiny links for your analysis and views of data. These are now stored for you so you may more easily find a previously run analysis. | ||
*3pm: reconfiguring the system | |||
*3:30pm: turn on web server | <br> | ||
*3:31pm: Nothing works | |||
*3:32pm: Start debugging | == CoGe v5 Deployment Process == | ||
*3:34pm: Get things working -- CoGe starts! | |||
Sept 24th, 2012 | |||
*9am: We shut down the website at 9am and started the final backup and freezing of existing data and analyses. | |||
*10am: Database is being replicated to the iPlant Data Store and copied to a backup server for processing and conversion to new database scheme. | |||
*11am: All web-code and libraries were backed up and new code deployed | |||
*12pm: updating database | |||
*1pm: copying database back to iRODS | |||
*2pm: copying database to coge server | |||
*3pm: reconfiguring the system | |||
*3:30pm: turn on web server | |||
*3:31pm: Nothing works | |||
*3:32pm: Start debugging | |||
*3:34pm: Get things working -- CoGe starts! | |||
*4:30pm: Most major problems found and corrected | *4:30pm: Most major problems found and corrected | ||
[[Image:Screen shot 2012-09-24 at 4.25.54 PM.png|thumb|center|400px]] | |||
== CoGe v5 Deployment == | |||
Sept 21st, 2012 | |||
CoGe v5 is planned for deployment on Sept. 24th. This new version of CoGe represents a massive revamping and extension of the user-data management system. | |||
Key features include: | |||
*Limited support for experimental data | |||
**This new system is funded (in part) by a grant from the Gordon and Betty Moore Foundation to add visualization support for epigenetics data for Arabidopsis. This is known as the EPIC-CoGe project: http://www.iplantcollaborative.org/learn/news/2012/05/24/iplant-ci-leveraged-development-epic-coge-browser | |||
*Ability to make lists and collections of data | |||
**Lists of experiments | |||
**Lists of genomes | |||
**Lists of features | |||
**Lists of lists | |||
*Enhancements for managing and sharing private data in CoGe | |||
*Logging user history so it is easier to find old analyses | |||
A key part to the migration to the new version of CoGe is preserving current private data in the system and assigning them to the appropriate owner. (We have done some major changes to the underlying metadata storage database for CoGe). Please let us know if you have lost access to your data and we will get that corrected right away. | |||
New features will be added that further integrate user specified lists into various tools in CoGe. E.g. auto-selecting a list of genomes for use in CoGeBlast instead of manually searching for all the genomes. | |||
Many thanks to CoGe Developer Matt Bomhoff for all the work on this new version. | |||
Please post any comments, suggestion, questions to CoGe's Forums (hosted by [http://iplantc.org iPlant]): https://forums.iplantcollaborative.org/viewforum.php?f=10 | |||
== Phaseolus vulgaris (common bean) v1 added to CoGe == | |||
Aug 23, 2012 | |||
Also located in CoGe [[ | Released from JGI/Phytozome, it is v1 of the common bean: http://genomevolution.org/CoGe/OrganismView.pl?oid=36223 | ||
[[Syntenic dotplots]] between it and soybean (Glycine max), [[Phaseolus vulgaris v. Glycine max]], clearly show that Phaseolus lacks the most recent [[Whole genome duplication]] in the Glycine lineage. | |||
== CoGe Paper published: '''Unleashing the genome of ''Brassica rapa'' ''' == | |||
July 31th, 2012 | |||
This Open Access paper provides a set of examples of how to analyze and compare the genome of '''Brassica rapa'''. Very useful for people wanting to learn how to use CoGe or how to maximize their use of the genome of '''Brassica rapa''': | |||
Open Access article in Frontiers of Plant Genetics and Genomics: http://www.frontiersin.org/plant_genetics_and_genomics/10.3389/fpls.2012.00172/abstract | |||
Also located in CoGe [[Tutorials]] sections. | |||
== Banana genome published == | == Banana genome published == | ||
| Line 87: | Line 103: | ||
*http://www.abc.net.au/science/articles/2012/07/12/3542969.htm | *http://www.abc.net.au/science/articles/2012/07/12/3542969.htm | ||
{| width="200" | {| width="200" cellspacing="1" cellpadding="1" border="1" | ||
|- | |- | ||
| [[ | | [[Image:R970403 10516168.jpeg|thumb|right|200px]] | ||
| [[ | | [[Image:Screen shot 2012-07-12 at 1.20.27 PM.png|thumb|right|200px]] | ||
|} | |} | ||
==Tomato genome published; the solanum hexaploidy investigated with CoGe== | == Tomato genome published; the solanum hexaploidy investigated with CoGe == | ||
May 31st, 2012 | |||
May 31st, 2012 | |||
The tomato genome was published in Nature earlier this week: | The tomato genome was published in Nature earlier this week: http://www.nature.com/nature/journal/v485/n7400/ | ||
However, the current version of the tomato genome has been in CoGe for the past year (thanks to an early release of the data from the tomato genome consortium). | However, the current version of the tomato genome has been in CoGe for the past year (thanks to an early release of the data from the tomato genome consortium). | ||
I've received a couple of emails inquiring about the Solanum specific hexaploidy, and this has been investigated with Haibao Tang. | I've received a couple of emails inquiring about the Solanum specific hexaploidy, and this has been investigated with Haibao Tang. Overall, these analyses support that the majority of the genome is derived from a tetraploidy, but there is evidence of some regions being triplicated (perhaps through a hexaploidy). | ||
These analyses are available: [[Tomato genome]] | These analyses are available: [[Tomato genome]] | ||
Please send us your thoughts or post them on the CoGe Forum: | Please send us your thoughts or post them on the CoGe Forum: | ||
# https://forums.iplantcollaborative.org/viewtopic.php?f=10&t=92 | |||
#https://forums.iplantcollaborative.org/viewtopic.php?f=10&t=92 | |||
== Video tutorial on how to use the iPlant Data Store to generate a quick-share link == | |||
May 29th, 2012 | |||
If you want to load a [[How to load a private genome into CoGe?|private genome into CoGe]], you need to send that genome to the CoGe team. This method makes it very easy for us to download your genome quickly! | |||
<youtube>CoHjYWSvrPA</youtube> | |||
== Dedicate CoGe Forum hosted at iPlant (part of the powered by iPlant program) == | |||
May 25th, 2012 | |||
iPlant has set up a dedicated forum for CoGe: https://forums.iplantcollaborative.org/viewforum.php?f=10 | |||
Please post any CoGe questions you have to here. | |||
== EPIC-CoGe Browser == | |||
May 24th 2012 | |||
May 24th 2012 | |||
News article about this project at iPlant: http://www.iplantcollaborative.org/learn/news/2012/05/24/iplant-ci-leveraged-development-epic-coge-browser | News article about this project at iPlant: http://www.iplantcollaborative.org/learn/news/2012/05/24/iplant-ci-leveraged-development-epic-coge-browser | ||
Overview of the Epic-CoGe Browser prototype system: | Overview of the Epic-CoGe Browser prototype system: | ||
<youtube>a_B8gd--XbQ</youtube> | <youtube>a_B8gd--XbQ</youtube> | ||
Try it: http://genomevolution.org/CoGe/GenomeView.pl?z=6&x=20000&dsgid=7043&chr=1 | Try it: http://genomevolution.org/CoGe/GenomeView.pl?z=6&x=20000&dsgid=7043&chr=1 | ||
WARNING: | WARNING: performance is a known issue! Some tiles in the browser may take a while to render (but are then cached). | ||
==2000 new genomes in CoGe== | == 2000 new genomes in CoGe == | ||
Apr. 30th 2012 | |||
The NCBI genome loader program was updated and run over the weekend. This resulting in about 2000 new genomes being loaded into CoGe. | |||
CoGe | == Unscheduled CoGe downtime == | ||
Apr. 25th 2012 | |||
CoGe was down/offline yesterday for two reasons: | |||
#One of iPlant's VMs was compromised and UITS (UA's IT group) shut off one of iPlant's subnets, which CoGe happens to use. This was due to a VM administered by a group collaborating with iPlant and not due to iPlant | |||
#Since CoGe was offline, when it came up, we decided to keep it offline for a while longer in order to updated the apache web server. After apache was updated, CoGe was brought online. Unfortunately, UITS detected a security vulnerability in the SSL implementation in the new update and shut CoGe off. This last part happened at the end of the day and we weren't able to coordinate with UITS to push a fix until this morning. | |||
While the CoGe team tries to keep as much uptime as possible, this type of downtime does happen once and a while. Our apologies to everyone whose work was interrupted or delayed due to this. | |||
== The algorithm, Last, added to [[SynMap]] == | |||
Mar. 28th 2012 | |||
Last (http://last.cbrc.jp/) has been added as a comparison algorithm in [[SynMap]]. Its performance is phenomenal! This is still under testing, so please let us know if you have any problems with it. Also, special thanks to Haibao Tang for writing the parallelized adapter for Last that is used by SynMap. Without this program, the integration would not have happened as quickly, easily, or smoothly. | |||
== CoGe used to decode the secret message in JCVI Synthetic Genome == | |||
Mar. 24th 2012 | |||
Mar. | |||
I heard that there was a secret message in the JCVI synthetic genome: Mycoplasma mycoides JCVI-syn1.0. Using CoGe, the DNA containing the secret messages was identified and decoded. Here is the walk-through of how this was done: [[Mycoplasma mycoides JCVI-syn1.0 Decoded]]. | |||
*'''WARNING:''' contains spoilers! | |||
*'''Note:''' this puzzle is nearly 2 years old. | |||
For those interested in doing the puzzle, this article has a good summary of the challenge: | |||
*Article from Sigularity Hub on the secret message: http://singularityhub.com/2010/05/24/venters-newest-synthetic-bacteria-has-secret-messages-coded-in-its-dna/ | |||
And you will probably need the original article (and the Supplementary Data): | |||
*Original Science Article on the genome: http://www.sciencemag.org/content/329/5987/52.abstract | |||
== CoGe Forums == | |||
Mar. 2nd 2012 | |||
iPlant has a forums site available: http://forums.iplantcollaborative.org | |||
CoGe, being part of the "Powered by iPlant" program, has a section on there for users to post questions about how to do various tasks, about CoGe in general, and provide suggestions. I'll be posting questions that are emailed to me there, but this will hopefully be a good place for people to ask questions, find answers, and help one another. | |||
= | Powered by iPlant Forum: https://forums.iplantcollaborative.org/viewforum.php?f=8 | ||
Update on genomes available from [http://www.phytozome.net Phytozome]. | The CoGe Forum: https://forums.iplantcollaborative.org/viewforum.php?f=10 | ||
== BlastN Bug == | |||
Feb. 11th 2012 | |||
Mike Freeling from UC Berkeley has found an interesting bug in BlastN where a relatively large blast hit (HSP) appears/disappears depending on the amount of sequence compared between Arabidopsis and Brassica. James Schnable from UC Berkeley further characterized this by identifying a comparison that differs in 1 nucleotide (over ~750) that causes this effect. You can see images of this blast error, characterization of the blast, an breakdown of parameters used here: [[GEvo Blastn Bug]] | |||
<br> | |||
== CoGe Server Migration == | |||
Feb. 4th 2012 | |||
CoGe's entire system has been migrated to the new server hosted by the [iplantcollaborative.org iPlant Collaborative]. This include | |||
*A new version of CoGe (v4) that includes: | |||
**Federation with iPlant's Authentication Services: [[How to get a CoGe account|read more]] | |||
**Rewritten backend for more modularization: [[Modularized CoGe|read more]] | |||
**User groups that can share and manage a restricted/private set of genomes: [[Groups|read more]] | |||
**Many updates to CoGe's tools | |||
**New genomes! | |||
*Migration of CoGePedia to new server | |||
*Migration of CoGe's tiny-url service (used to construct URLs that can be used to regenerated analysis -- mainly by [[GEvo]] and [[SynMap]]) | |||
*Update to DNS for: | |||
**http://genomevolution.org | |||
**http://genomeevolution.org | |||
**http://genomevolution.com | |||
Please [[Contact Page|contact us]] if you come across any problems! | |||
== Exciting new plant genomes in CoGe == | |||
Feb. 3rd 2012 | |||
Update on genomes available from [http://www.phytozome.net Phytozome]. | |||
The genomes of | The genomes of | ||
*[[Sequenced plant genomes# | |||
*[[Sequenced plant genomes# | *[[Sequenced plant genomes#Common_bean|common bean]] (a crucial staple food of grad students everywhere) | ||
**Syntenic dotplot: [[Capsella rubella - Arabidopsis lyrata]] | *[[Sequenced plant genomes#Capsella_rubella|capsella]] (the close relative of arabidopsis, not [http://www.youtube.com/watch?v=8J1iDbLnEAg the song by I:Scintilla]) | ||
*Linum usitatissimum (common flax; linseed): http://genomevolution.org/CoGe/OrganismView.pl?oid=36226 | **Syntenic dotplot: [[Capsella rubella - Arabidopsis lyrata]] | ||
**Syntenic dotplot: | *Linum usitatissimum (common flax; linseed): http://genomevolution.org/CoGe/OrganismView.pl?oid=36226 | ||
**Syntenic dotplot: [[Flax -Poplar]] | |||
*Gossypium raimondii (cotton): http://genomevolution.org/CoGe/OrganismView.pl?oid=36239 | *Gossypium raimondii (cotton): http://genomevolution.org/CoGe/OrganismView.pl?oid=36239 | ||
have both been added to iPlant CoGe. Head over and check them out. | have both been added to iPlant CoGe. Head over and check them out. <-- But remember these genomes are protected by Fort Lauderdale for the next twelve months or until you see the genome paper. | ||
<br> | |||
Are we missing plant genomes you'd like to be studying? [[Contact Page|Let us know!]]. | |||
== iPlant User Management System Update == | |||
Dec. 18th 2011 | |||
The [[Data security model]] of CoGe has been updated. This includes creating CoGe [[Groups]] which permits the creation of user groups. These user groups may access a private set of genomes that is not accessible to other users of CoGe. | |||
To use this, you will need to create an account with iPlant in order to be a registered [[CoGe user]]: | |||
== Major CoGe Update (version 4) == | |||
Dec. 4th 2011 | |||
Work is nearing completion for a new version of CoGe. While there are many minor improvements, additions, and changes to the tools, the major improvements are on the backend of the system including: | |||
*New server hosted by iPlant: This means that the primary CoGe server will be located at the University of Arizona, Tucson | |||
*New server hosted by iPlant: | **Vastly expanded storage to hold even more genomes | ||
**Vastly expanded storage to hold even more genomes | **Enables the storage of metagenomes (as those datasets can be quite large) | ||
**Enables the storage of metagenomes (as those datasets can be quite large) | *Modularized installation and centralized configuration: permits the rapid deployment of custom versions of CoGe (for those that may want a version of CoGe specific to their group of organisms) | ||
*Modularized installation and centralized configuration: permits the rapid deployment of custom versions of CoGe (for those that may want a version of CoGe specific to their group of organisms) | *Federation with iPlant's authentication system: | ||
*Federation with iPlant's authentication system: | **People will iPlant login credentials can log into CoGe as a registered user. | ||
** People will iPlant login credentials can log into CoGe as a registered user. | **Will enable the creation of personal data in CoGe | ||
** Will enable the creation of personal data in CoGe | **Will enable more customization and saving of preferences for various tools in CoGe | ||
** Will enable more customization and saving of preferences for various tools in CoGe | **Will enable users to save particular analyses and datasets within CoGe | ||
** Will enable users to save particular analyses and datasets within CoGe | **Will enable import and export of data from CoGe to people's iPlant Data Store accounts | ||
** Will enable import and export of data from CoGe to people's iPlant Data Store accounts | |||
*Enhanced data security model: | *Enhanced data security model: | ||
** Will enable unpublished data to be restricted to a user or a group of users | **Will enable unpublished data to be restricted to a user or a group of users | ||
Please come test the new CoGe: http://coge.iplantcollaborative.org and send [mailto:elyons.coge@gmail.com Eric Lyons] any problems you come across. | |||
Since the holidays are coming and usage of CoGe tends to decrease, hopefully any bugs won't affect too many people while they are fixed. The migration of the domain names registered to CoGe will change once the server has been reasonably tested. Other CoGe services will migrate after that (e.g. this wiki). | |||
CoGe domain names: | |||
*http://genomevolution.org | |||
*http://genomevolution.org | *http://genomeevolution.org | ||
*http://genomeevolution.org | |||
*http://genomevolution.com | *http://genomevolution.com | ||
==Pigeon-pea genome (Cajunus cajan) | == Pigeon-pea genome (Cajunus cajan) has been added to CoGe == | ||
Nov. 29th, 2011 | |||
Nov. 29th, 2011 | |||
The [http://www.icrisat.org/gt-bt/iipg/Home.html International Initiative for Pigeonpea Genomics] has released the pigeon pea genome. | |||
The | The pigeon-pea genomes may be accessed in CoGe: http://genomevolution.org/CoGe/OrganismView.pl?oid=34028 | ||
Please see this link for a syntenic dotplot between pigeonpea and medicago: http://genomevolution.org/r/49ua This syntenic dotplot has the syntenic gene pairs' evolution distance colored to differentiate orthologous and out-paralogous syntenic regions. | |||
<br> | |||
== NCBI Genome Update: Over a thousand new genomes available in CoGe == | |||
Nov. 28th, 2011 | |||
Nov. 28th, 2011 | |||
The NCBI genome loading program for CoGe has been updated as is currently adding thousands of genomes from NCBI. | The NCBI genome loading program for CoGe has been updated as is currently adding thousands of genomes from NCBI. Keeping CoGe current with all of the genomes at NCBI has been a challenge as their underlying data model for storing and organizing genomes evolves. The new program crawls all of NCBI's BioProjects searching for those with genomes and associated sequence. Prior to this data load there were approximately 12,100 genomes from 10,600 organisms. Approximately 40% of NCBI's BioProjects have been crawled and the current genome stats are: | ||
Organisms: 12,093 | Organisms: 12,093 | ||
Genomes: 13,969 | |||
Nucleotides: 305,480,720,992 | |||
Genomic Features: 99,814,749 | |||
Annotations: 224,582,292 | |||
For those that are curious, CoGe has maintained a MySQL DB transaction rate of 2000-3000 per second (majority writes/inserts) for the past 24 hours, thanks in no small part to its SSD configuration. | |||
'''Note:''' After more performance monitoring, peak DB transactions top 9000 per second during heavy use from the genome loading programs and website activity. | |||
== Optical fun with CoGe == | |||
Nov. 22nd, 2011 | |||
[[Image:DNA orbit animated small-side.gif]] | |||
Which direction does the DNA spin? Depending on how your mind is interpreting the dark and light colored dots of the DNA molecule as being "near" or "far", the helix can spin in both directions. | |||
Thanks to Don McCarty for pointing this out. | |||
== Lamprey, Anole, and Frog genomes added/updated to CoGe == | |||
Nov. 19th, 2011 | |||
[www.ensembl.org Ensembl] version 64 genomes of Lamprey, Anole, and Frog have been added to CoGe: | |||
Petromyzon marinus (lamprey): http://genomevolution.org/CoGe/OrganismView.pl?oid=30737 Xenopus (Silurana) tropicalis (western clawed frog): http://genomevolution.org/CoGe/OrganismView.pl?oid=33964 Anolis carolinensis (green anole): http://genomevolution.org/CoGe/OrganismView.pl?oid=33828 | |||
Both the unmasked and [[Masked]] versions of the genomes are available. For an example [[Syntenic dotplot]] between Xenopus and Tetraodon (pufferfish), please see: http://genomevolution.org/r/48w9 | |||
This dotplot uses the [[Syntenic path assembly]] to order and orient the contigs of Xenopus to the well assembled genome of Tetraodon (Frog versus Pufferfish): http://genomevolution.org/r/48w9 | |||
The CoGePedia [[Sequenced plant genomes]] page has been updated with the latest published genomes, including the just published genomes of both [[Sequenced plant genomes#Cannabis|pot]] and [[Sequenced plant genomes# | This dotplot uses the [[Syntenic path assembly]] to order and orient the contigs of Xenopus and Anolis (Frog V Green Lizard): http://genomevolution.org/r/48zk | ||
*[[Plant Genome Statistics|Plant Genome Papers]] lists the papers describing every published plant genome, when and where it was published, and how much attention (in the form of citations) the various genomes have attracted so far. | |||
Thanks to Bill Spollen for requesting these genomes. | |||
== Updated and New Plant Genome Resources == | |||
Nov. 10th 2011 | |||
The CoGePedia [[Sequenced plant genomes]] page has been updated with the latest published genomes, including the just published genomes of both [[Sequenced plant genomes#Cannabis|pot]] and [[Sequenced plant genomes#Pidgeon_Pea|pidgeon pea]]! In addition, we have added two new pages that may be of interest to those who (like me) are constantly having to pull together introduction sections and can't remember what the right citation for well known genomic information is: | |||
*[[Plant Genome Statistics|Plant Genome Papers]] lists the papers describing every published plant genome, when and where it was published, and how much attention (in the form of citations) the various genomes have attracted so far. | |||
*[[Plant paleopolyploidy]] is a list of known ancient whole genome duplications among the various plant species with sequenced genomes including information on when and how the whole genome duplications were discovered. | *[[Plant paleopolyploidy]] is a list of known ancient whole genome duplications among the various plant species with sequenced genomes including information on when and how the whole genome duplications were discovered. | ||
Both pages are clearly works in progress so please continue to contact us if we've missed genomes, whole genome duplications, or citations which should be on the list. | Both pages are clearly works in progress so please continue to contact us if we've missed genomes, whole genome duplications, or citations which should be on the list. | ||
== Main CoGe Database is down == | |||
Nov. 3rd 2011 | |||
''7:00 (PCT USA) 14:00 (GMT)'' | |||
' | Last night I ran a repair table on the main database for CoGe. This apparently ran into some problems and failed. I am currently hunting down the problem, and the main CoGe site is currently off-line. Technically, the tools are all available, but some of them are not working. The problem appears to be located in the "locations" table of the [CoGe database]. This table records the locations for all of CoGe's [[Genomic features]]. For anyone that needs to get some work done with CoGe, they are welcome to use the development server hosted at: | ||
http://coge.iplantcollaborative.org | |||
This version of CoGe has been under development to federate CoGe's user authentication system with the authentication system provided by the iPlant Collaborative. As such, there has been many code changes dealing with registered users and accessing restricted/private genomes. These changes are NOT fully tested and may cause some problems. Also, the development server is using an out-of-date version of the main CoGe database (though most of the genomes should be there). If you use the development server and run into any of these problems, please feel free to send [mailto:elyons.coge@gmail.com Eric Lyons] an email. I'd appreciate the reporting of any bugs as well as your patience with the current situation. | |||
In case of catastrophic failure of the main database, please know that in addition to the development server, there is a full backup of the main CoGe database. These are generated weekly. | |||
Also, thanks to Ben Field for notifying me of the problem. I deeply appreciate the help of community members in alerting me to problems with the site as well as suggestions for making it better. | |||
''Update: 8:00am'' | |||
*Another "repair table" is being run on the main CoGe Database. | |||
*Another "repair table" is being run on the main CoGe Database. | *Backup database is being restored on the dev server for CoGe (coge.iplantcollaborative.org). Once this is up and running, I'll point the main CoGe site to use this database and database server in case the main database has not yet been repaired. | ||
*Backup database is being restored on the dev server for CoGe (coge.iplantcollaborative.org). | |||
''Update: 9:30am'' | |||
*backup coge database has been deployed to CoGe development server, currently undergoing "optimization" (want to avoid whatever happened to the main database) | *backup coge database has been deployed to CoGe development server, currently undergoing "optimization" (want to avoid whatever happened to the main database) | ||
''Update: 5pm'' | ''Update: 5pm'' | ||
*main coge database has been repaired. Warning and update messages taken down from the website. Let me know if anyone has any problems. | |||
== CoGe Tutorial Published in [http://www.maydica.org/ Maydica:] == | |||
Oct. 24th 2011 | |||
A comprehensive open-access tutorial on using CoGe has been published in Maydica: http://www.maydica.org/articles/56_183.pdf | |||
<br> '''Abstract:''' | |||
Of all the major plant groups, the grasses, with the complete genomes of five species, are the best positioned to take advantage of comparative genomics to obtain insight into functional genetic elements. Of all the grasses, maize is the best characterized in terms of genetics, development, and evolution. We provide several examples of how the web-based comparative genomics system CoGe may be used to aid in the interpretation of the maize genome sequence. These examples include verifying gene models, identifying differences between genome as- semblies, identifying conserved non-coding sequences, identifying syntenic regions between species and poly- ploidies, and identifying homeologs within maize and orthologs between maize and other grass genomes. In addition, a comprehensive list of orthologous gene sets is provided between maize and Sorghum, foxtail millet, rice, and Brachypodium. | |||
While the article focuses on the maize genome as its primary genome, the methods are applicable to any genome. | <br> While the article focuses on the maize genome as its primary genome, the methods are applicable to any genome. | ||
== Correction to the [[ | == Correction to the [[Classical Maize Genes#The_Table|Classical Maize Gene and Syntelog List]] == | ||
Sept. 29th 2011 | |||
The following genes are now assigned as being not present in the B73: | Phil Stinard identified an error in incorrectly assigning classical maize genes as being present in B73. Thanks to Mary Schaeffer for passing along this information and James Schnable for correcting these in the [[Classical Maize Genes#The_Table|Classical Maize Gene and Syntelog List]]. | ||
*S | |||
*lc1 | The following genes are now assigned as being not present in the B73: | ||
*sn1 | |||
*S | |||
*lc1 | |||
*sn1 | |||
*hopi1 | *hopi1 | ||
== New options in [[SynMap]] == | == New options in [[SynMap]] == | ||
Sept. 12th, 2011 | |||
Sept. 12th, 2011 | |||
There are a couple of new options available in SynMap: | |||
'''Force dotplot to be a square:''' You can find this option under the "Display Options" Tab with the line "Dotplot axes relations". | |||
'''SVG Version of the Dotplot:''' There will be a new file, "SVG Version of the Syntenic Dotplot" to download in the "Links and Downloads" section of the results. This file will only appear if some form of synonymous rates are calculated and visualized (available under the "Analysis Options" tab"). | |||
<br> Thanks to James Schnable for creating the SVG program for SynMap! | |||
== Potato genome added to CoGe == | |||
Sept. 3rd, 2011 | |||
Genome published: http://www.nature.com/nature/journal/v475/n7355/full/nature10158.html | |||
The genome added was doubled the monoploid S. tuberosum Group Phureja clone DM1-3 516R44 (DM): | |||
#unmasked: http://genomevolution.org/CoGe/OrganismView.pl?dsgid=12277 | |||
#masked: http://genomevolution.org/CoGe/OrganismView.pl?dsgid=12278 | |||
Please note: this version of the genome does not have annotations available. | |||
Thanks to Will Spooner for the notification! | |||
== Brassica rapa genome added to CoGe == | |||
Sept. 3rd, 2011 | |||
Genome published: http://www.nature.com/ng/journal/vaop/ncurrent/full/ng.919.html#/group-1 | |||
Sequenced by: [http://www.genomics.cn BGI] | |||
Brassica rapa has had a hexaploidy event subsequent to the most recent tetraploidy event in the Arabidopsis lineage. | |||
Thanks to Will Spooner for the notification! | |||
== Cannabis sativa [[Pseudoassembled]] genome added to CoGe == | |||
Aug. 23rd, 2011 | |||
[[SynMap]] has the option to assembled one genome against another using syntenic. Such [[Syntneic path assemblies]] may be used to create a [[Pseudoassembly]] of a genome when only a contig level assembly exists. [[SynMap]] makes generating these [[Pseudoassemblies]] easy to do. Such a [[Pseudoassembly]] of the 175,000 Cannabis sativa genome was performed against the peach genome ([[Why peach and grape genomes are peachy!|read here to learn why peach was chosen]]). This pseudoassembly was reloaded back into CoGe and permits using CoGe's tools to compare the Cannabis genome at multiple levels of resolution. | |||
[[ | '''To see this example:''' [[Cannabis sativa cultivar Chemdawg (marijuana)]] | ||
[[Pseudoassemblies]] may be quite useful as more genomes are sequences on the cheap. Such sequencing project yield low-quality draft genomes that are usually assembled into several tens of thousands of contigs, and pseudoassemblies permit the rapid generation of large sequences that are easier to use in comparative genomic analyses. | |||
== Cannabis sativa cultivar Chemdawg (marijuana) added to CoGe == | |||
Aug. 22nd, 2011 | |||
Aug. 22nd, 2011 | |||
The genome of the extremophile Cannabis sativa cultivar Chemdawg (marijuana) has been added to CoGe: | The genome of the extremophile Cannabis sativa cultivar Chemdawg (marijuana) has been added to CoGe: http://genomevolution.org/CoGe/OrganismView.pl?oid=33804 | ||
This genome was sequenced by [http://www.medicinalgenomics.com/ Medicinal Genomics] (located in the Netherlands). | This genome was sequenced by [http://www.medicinalgenomics.com/ Medicinal Genomics] (located in the Netherlands). It was sequenced with one lane of the Illumina HiSeq (2x100) platform and assembled with CLCbio’s workbench. Additional information about the assembly and genome may be found: http://www.medicinalgenomics.com/the-c-sativa-genome/ | ||
'''You can access Cannabis in CoGe:''' http://genomevolution.org/CoGe/OrganismView.pl?oid=33804 | '''You can access Cannabis in CoGe:''' http://genomevolution.org/CoGe/OrganismView.pl?oid=33804 | ||
Cannabis is a member of the plant order Rosales. | Cannabis is a member of the plant order Rosales. Of sequenced genomes in that order, the peach genome is a [[Why peach and grape genomes are peachy!|fantastic comparator]]. The reason for this is due to its high-quality sequence and assembly, and its genomic evolutionary history that does not contain any whole genome duplication event subsequent to the [[Eudicot paleohexaploidy]] shared by nearly all dicots (at least the eurosids and the astrids). As such, its genome structure is probably very similar to the common ancestor of order Rosales, and perhaps the eudicots as a whole. This likely ancestral state of the peach genome makes it quite suitable for generating a [[Pseudoassembly]] of highly fractured, low quality genome assemblies such as this Cannabis genome. CoGe's tool [[SynMap]] has an algorithm to tile contigs along any other "reference" genome in CoGe. | ||
The [[Syntenic path assembly]] of Cannabis to the peach genome may be viewed: http://genomevolution.org/wiki/index.php/Syntenic_path_assembly#Cannabis_sativa_.28marijuana.29_v._Prunus_persica_.28peach.29 | The [[Syntenic path assembly]] of Cannabis to the peach genome may be viewed: http://genomevolution.org/wiki/index.php/Syntenic_path_assembly#Cannabis_sativa_.28marijuana.29_v._Prunus_persica_.28peach.29 | ||
This shows the Cannabis genome sequence contains nearly the entire gene content of Peach. | This shows the Cannabis genome sequence contains nearly the entire gene content of Peach. | ||
== Eutrema parvulum (Thellungiella parvula) added to CoGe == | == Eutrema parvulum (Thellungiella parvula) added to CoGe == | ||
Aug. 17th, 2011 | |||
The genome of the extremophile crucifer Eutrema parvulum (Thellungiella parvula) has been added to CoGe: http://genomevolution.org/CoGe/OrganismView.pl?dsgid=12242 | |||
You can read about this genome in this Nature Genetics Letter: http://www.nature.com/ng/journal/vaop/ncurrent/full/ng.889.html | |||
For a syntenic dotplot between it and Arabidopsis thaliana, please this [[SynMap]] anlaysis: http://genomevolution.org/r/3ws0 | |||
<br> | |||
Version | == New Version of Setaria italica (foxtail millet) added to CoGe == | ||
Aug. 16th, 2011 | |||
Version 2.1 of Setaria italica has been added to CoGe. This genome was obtained from JGI/phytozome: http://phytozome.net | |||
= | Unmasked version: http://genomevolution.org/CoGe/OrganismView.pl?dsgid=12240 Masked version: http://genomevolution.org/CoGe/OrganismView.pl?dsgid=12241 | ||
Thanks to Gina Turco for the request. | |||
This | == New Version of Fragaria vesca (woodland strawberry) added to CoGe. This time with gene models! == | ||
Aug. 11th, 2011 | |||
Version 1.1 of Fragaria vesca (woodland strawberry) has been added to CoGe http://genomevolution.org/CoGe/OrganismView.pl?dsgid=12186 . | |||
This version contains gene models with permits more fun with syntenic dotplots: http://genomevolution.org/r/3wdb | |||
This dotplot is strawberry versus peach. Besides from be a great summer fruit salad, this dotplot colors syntenic gene pairs based on their synonymous mutation values. From it, it is easy to see neither genome has had an independent whole genome duplication since the [[Eudicot paleohexaploidy]] event. | |||
Thanks to Aaron Liston for requesting this genome. | |||
== Daphnia pulex (common water flea) added == | |||
Aug. 3rd, 2011 You can get all your water flea genomics here: http://genomevolution.org/CoGe/OrganismView.pl?oid=33760 | |||
Thanks to Mike Freeling for the request. | |||
== Several bugs fixed as a result of the code update == | |||
July 29th, 2011 | |||
July | |||
CoGe | Additional bugs were squashed today due to the major code update to CoGe's internal services. Part of the update included further modularization of the web-services from backend services. A few programs the ancillary support programs for CoGe's web-services were not correctly being passed the base configuration file for a given web-deployment and were therefore crashing. This has been corrected, but please email [mailto:elyons.coge@gmail.com Eric Lyons] if any problems are encountered. | ||
== Update to [[GenomeList]] == | |||
July 29th, 2011 | |||
GenomeList has been updated to: | |||
#include a link back to GenomeList for selected genomes. This is useful if a broad selection of genomes was made and needs to be refined. | |||
#include a link to easily download a fasta file for a given genome | |||
#include a link to coge_gff to generate a gff file of all [[Genomic features]] and annotation in a genome | |||
#include a [[TinyURL]] link to regenerate the genome list. This link is found at the top of the genome list. | |||
Example GenomeList link: http://genomevolution.org/r/3v8n | |||
== Major code update to CoGe == | |||
July 27th, 2011 | |||
CoGe has undergone a major update of its web-based system today. A few bug fixes and feature enhancements mixed in, with the major one being the addition of GenomeList for creating a list of genomes, getting an overview of their genomic content, and then sending the list to other tools (e.g. CoGeBlast). | |||
Behind the scenes was a further modularization of the web-interface from the backend support services and modules. The primary reason for this is to enable to creation of multiple CoGe installations. There has been a few requests by people for a clade/group of organisms specific installation of CoGe. With [http://iplantc.org iPlant's cyberinfrastructure] support, this should be possible (providing the code-base supports it). | |||
There were some sticking points this morning migrating server specific changes from the iPlant development server to the main CoGe server, but hopefully this didn't affect too many people. ''However, there is a high-likelihood of additional bugs in the system that I failed to catch!'' Please email [mailto:elyons.coge@gmail.com Eric Lyons] if you find any problem. | |||
Otherwise, we are hoping to make a full migration to iPlant's resources in the near future. iPlant's coge server is being upgraded with some additional attached storage for continual growth of the platform. | |||
== [http://qatar-weill.cornell.edu/ Weill's] [http://qatar-weill.cornell.edu/research/datepalmGenome/download.html Date Palm] genome version 3 has been added to CoGe == | |||
July 4th, 2011 | |||
You can find its genome | You can find its genome in [[OrganismView]]: http://genomevolution.org/CoGe/OrganismView.pl?dsgid=11942 | ||
And a [[Syntenic path assembly]] to rice here: http://genomevolution.org/r/3ox8 | |||
This is a very rough genome (50,000+ contigs; the largest is 470KB; 13 larger than 300KB). However, the syntenic path assembly in [[SynMap]] with the option to remove any contig that doesn't have a syntenic signal makes identifying sytnenic regions a breeze (see the above link). | |||
See this example of micro-synteny as seen in GEvo: http://genomevolution.org/r/3oxa | |||
Thanks to: Haibao Tang, Devin O'Connor, and Jim Leebens-Mack for requesting this genome. | |||
July 14th, 2011 | |||
The masked version of the Palm genome has been created and added to CoGe: http://genomevolution.org/CoGe/OrganismView.pl?dsgid=11947 | |||
Thanks to Haibao Tang for providing the masking procedure. | |||
== [http://www.jgi.doe.gov/genome-projects/ JGI's] Eucalyptus grandis BRASUZ1 has been added to CoGe == | |||
You can find its genome here: http://genomevolution.org/CoGe/OrganismView.pl?oid=33537 (masked and unmasked sequence) | |||
With a comparison to the peach genome, Eucalyptus looks to have had its own whole genome duplication subsequent to the [[Eudicot paleohexaploidy]]: http://genomevolution.org/r/3ol1 | |||
The entire set of sequences and associated annotations for Coccidioides has been added to CoGe. | Thanks to Josquin Tibbits for recommending this genome! | ||
== Arabidopsis thaliana resequenced genomes (C24, Bur-0, Kro-0, Ler-1) from 1001genomes.org has been added to CoGe == | |||
June 30th, 2011 | |||
The "High Quality" sequences generated by the [http://1001genomes.org/projects/MPISchneeberger2011/index.html 1001genomes] project for the resequencing of several arabidopsis strains has been added to CoGe. This includes: | |||
*[http://synteny.cnr.berkeley.edu/CoGe/OrganismView.pl?dsgid=11934 Bur-0] | |||
*[http://synteny.cnr.berkeley.edu/CoGe/OrganismView.pl?dsgid=11933 C24] | |||
*[http://synteny.cnr.berkeley.edu/CoGe/OrganismView.pl?dsgid=11935 Kro-0] | |||
*[http://synteny.cnr.berkeley.edu/CoGe/OrganismView.pl?dsgid=11937 Ler-1] | |||
While these genomes contain many contigs, CoGe's [http://genomevolution.org/wiki/index.php/Syntenic_path_assembly#Arabidopsi_ecotypes:__Columbia_versus_Landsberg_erecta Syntenic path assembly] algorithm can arrange and orient them against the reference genome Col-0: http://genomevolution.org/r/3okf | |||
Thanks to Maggie Woodhouse for this suggestion! | |||
== OrganismView's Feature List display updated == | |||
June 22nd, 2011 | |||
OrganismView has a minor update for where the lists of [[Genomic features]] are displayed. The old version would display the summary list of genomic features below all the information panels. This would mean that each time a summary list was generated, it would replace the prior one. For example, if you retrieved the list first for the entire genome and second for a particular chromosome. Now, each information panel's genomic feature list appears to the right of the information summary. This allows the entire genome's feature list to be display simultaneously with the chromosome's feature list. | |||
<br> | |||
== [http://www.broadinstitute.org/ Broad Institute's] Coccidioides group Database added to CoGe == | |||
June 21st, 2011 | |||
The entire set of sequences and associated annotations for Coccidioides has been added to CoGe. These soil fungi are pathogenic and can cause coccidioidomycosis, aka valley fever, in humans. The original data may be obtained from: http://www.broadinstitute.org/annotation/genome/coccidioides_group/MultiHome.html | |||
And accessed through [[OrganismView]]: http://genomevolution.org/CoGe/OrganismView.pl?org_desc=Coccidioides | And accessed through [[OrganismView]]: http://genomevolution.org/CoGe/OrganismView.pl?org_desc=Coccidioides | ||
Thanks to Marc Orbach for suggesting and locating these genomes. | Thanks to Marc Orbach for suggesting and locating these genomes. | ||
== UC Berkeley Data Center Back Online == | |||
June 12th, 2011 | |||
The UC Berkeley Data Center power upgrade went smoothly. CoGe has booted up and is back online. | |||
Thanks to: | |||
*James Schnable for being on duty to bring CoGe down and back up. | |||
* James Schnable for being on duty to bring CoGe down and back up. | *The entire team at the UC Berkeley Data Center for completing such a complicated upgrade to their Center and for continually updating their clients as to the progress of the operation. | ||
* The entire team at the UC Berkeley Data Center for completing such a complicated upgrade to their Center and for continually updating their clients as to the progress of the operation. | |||
== CoGe Downtime June 12th, 2011 == | == CoGe Downtime June 12th, 2011 == | ||
| Line 570: | Line 637: | ||
''As part of this effort, the replacement of some core components of the data center’s power infrastructure is required. For safety reasons, a full power outage to the data center is scheduled for Sunday, June 12, 2011, from 7:00 am to 3:00 pm. The data center will rely entirely on outside air, rather than air conditioning, to provide cooling for the duration of this period. A minimal number of systems with broad campus impact, including CalMail, CalAgenda, and the campus home page, will be provided with temporary power during this outage. In the unlikely event that the data center air temperature exceeds a level appropriate for the safe operation of equipment, some of these systems may need to be shut down as well.'' | ''As part of this effort, the replacement of some core components of the data center’s power infrastructure is required. For safety reasons, a full power outage to the data center is scheduled for Sunday, June 12, 2011, from 7:00 am to 3:00 pm. The data center will rely entirely on outside air, rather than air conditioning, to provide cooling for the duration of this period. A minimal number of systems with broad campus impact, including CalMail, CalAgenda, and the campus home page, will be provided with temporary power during this outage. In the unlikely event that the data center air temperature exceeds a level appropriate for the safe operation of equipment, some of these systems may need to be shut down as well.'' | ||
''The list of widely used systems that are intended to remain available is below. This list is still being finalized, so additional systems may be added as campus needs require. This list will not include systems for which departments have made separate arrangements.'' | ''The list of widely used systems that are intended to remain available is below. This list is still being finalized, so additional systems may be added as campus needs require. This list will not include systems for which departments have made separate arrangements.'' | ||
== Citrus genomes added == | |||
May 6th 2011 | |||
The genomes of: | |||
*Citrus clementina (Clementine mandarin): http://genomevolution.org/CoGe/OrganismView.pl?oid=33274 | |||
*Citrus sinensis (citrus, Sweet orange): http://genomevolution.org/CoGe/OrganismView.pl?oid=33273 | |||
Have been added to CoGe. These were sequenced by [http://www.jgi.doe.gov/ JGI]. | |||
A quick syntenic analysis of sinensis to peach shows that it appears to have no subsequent whole genome duplication event to the eurosid [[Paleohexaploidy]]: http://genomevolution.org/r/2zdv | |||
== Sequenced Plant Genome Phylogeny Update == | |||
May 6th 2011 | |||
James Schnable has updated the phylogeny of angiosperms for [[Sequenced plant genomes|sequenced plant genomes]]. | |||
== CoGe Workshop at Berkeley == | |||
Apr. 19th 2011 | |||
Apr. 19th 2011 | |||
Here is the outline/syllabus of the workshop help at Berkeley hosted by the [http://iplantcollaborative.org iPlant Collaborative], the [http://pmb.berkeley.edu Department of Plant and Microbial Biology], [http://qb3.berkeley.edu/qb3/corefacilities.cfm QB3-CGRL (Computational Genomics Resource Laboratory)], [http://www.pgec.usda.gov/ ARS-Plant Gene Expression Center], and the [http://microscopy.berkeley.edu/~freeling/ Freeling lab]: | Here is the outline/syllabus of the workshop help at Berkeley hosted by the [http://iplantcollaborative.org iPlant Collaborative], the [http://pmb.berkeley.edu Department of Plant and Microbial Biology], [http://qb3.berkeley.edu/qb3/corefacilities.cfm QB3-CGRL (Computational Genomics Resource Laboratory)], [http://www.pgec.usda.gov/ ARS-Plant Gene Expression Center], and the [http://microscopy.berkeley.edu/~freeling/ Freeling lab]: [[2011 Berkeley Workshop]] | ||
This outline contains links to specific analyses used in the workshop. | This outline contains links to specific analyses used in the workshop. | ||
==Horizontal Genome Transfer== | == Horizontal Genome Transfer == | ||
Mar. 31st 2011 | |||
Here is a fun example of a mitochondria genome being inserted into a plant chromosome: Horizontal transfer of mitochondria genome: [[Horizontal transfer of mitochondria genome]] | |||
== Second "Run GEvo Analysis!" button added to [[GEvo]] == | |||
Mar. 29th 2011 | |||
For those times when scrolling to the top of the screen to find the "Run GEvo Analysis!" button is too much work, a second button has been added at the bottom of the configuration box. This is quite useful when comparing >6 genomic regions. | |||
Thanks to | Thanks to David Braun for this suggestion! | ||
== | == Bug Fix in FeatView == | ||
Mar. | Mar. 29th 2011 | ||
Thanks to Damon Lisch for pointing out a bug in FeatView that was exposed by Firefox v4. This bug was also affecting Google Chrome (but not Safari). Please let [mailto:elyons.coge@gmail.com Eric Lyons] know of any problems you have running Firefox v4 (or other problems in general). | |||
== New tutorial for performing genomic rearrangement analyses == | |||
Mar. 11th 2011 | |||
A new tutorial has been written for showing how to figure [[SynMap]] to generate a link to [http://grimm.ucsd.edu/GRIMM/ GRIMM] (by Glenn Tesler, University of California, San Diego) for performing genomic rearrangement analysis. | |||
Tutorial: [[Tutorials#How_to_perform_a_genomic_rearrangement_analysis|How to perform a genomic rearrangement analysis]] | |||
== [[SynMap]] now has support for BlastP == | |||
Mar. 7th 2011 | |||
You can now select to compare protein sequences between genomes with annotated protein coding features ([[CDS]]). | |||
Thanks to Angelique D'Hont for the suggestion. | |||
== Cochliobolus heterostrophus C5 from JGI loaded into CoGe == | |||
Mar. 2nd 2011 | |||
You can find Cochliobolus heterostrophus C5 in [[OrganismView]]: http://genomevolution.org/CoGe/OrganismView.pl?dsgid=11258 | |||
Both masked (by JGI) and unmasked version of the genome are available. | |||
For a syntenic dotplot between C. heterostrophus to Pyrenophora tritici-repentis strain Pt-1C-BFP (the closest relative I could find in CoGe) please follow: http://genomevolution.org/r/2m0n | |||
This is a neat syntenic dotplot showing extensive synteny and intrachromosomeal rearrangements (though these are both contig level assemblies). | |||
Thanks to Daniel Lawrence for request. | |||
Thanks to: | == Sort chromosomes by name in SynMap == | ||
*Angélique D'Hont from CIRAD | |||
Feb. 26th 2011 | |||
After a couple of requests, SynMap now has an option to sort chromosomes by name instead of by size. You can read [[SynMap#How_do_I_sort_the_chromosomes_by_name_instead_of_by_size.3F|how to set this option '''here''']]. | |||
Thanks to: | |||
*Angélique D'Hont from CIRAD | |||
*James Schnable from UC Berkeley | *James Schnable from UC Berkeley | ||
for this suggestion. | |||
== How to load genomes into CoGe == | |||
Feb. 22nd 2011 | |||
If you have a CoGe installation, access to the main CoGe server, or just curious to know what is needed to load a genome into CoGe, here is a page on [[How to load genomes into CoGe]]. This is all run from the command line, and when CoGe's user permission data management system matures, this procedure will be made available via the web. | |||
<br> | |||
== Giant Panda genome loaded into CoGe == | |||
Feb. 19th 2011 | |||
You can see the genome in CoGe at: http://genomevolution.org/CoGe/OrganismView.pl?dsgid=11227 | |||
This was one of the [http://www.nature.com/nature/journal/v463/n7279/full/nature08696.html first big genomes] sequenced using only [[Next Generation Sequencing Technology]] and assembled [[De novo]]. As a result, the assembly is rather poor compared to a fully assembled genome like [http://www.nature.com/nature/journal/v438/n7069/full/nature04338.html the dog genome]. However, through comparative genomics with [[SynMap]], identifying syntenic regions and determining that nearly full coverage was obtained is as easy as a few mouse clicks: [[Syntenic path assembly#Carnivora:_Giant_Panda_.28WGS_Assembly.29_to_Dog_.28reference_genome.29|syntenic path assembly of the WGS panda genome to the fully sequenced dog genome]]. This will be quite useful as more and more large genomes are sequenced using these techniques (fast, cheap, and still very useful!) | |||
== First Metagenome loaded into CoGe == | |||
Feb. 19th 2011 | |||
[ | Technically, there is no reason why CoGe can't store metagenomes. Its [[CoGe Database|core data model]] stores a collection of sequences that, thus far, has been organized into a genome, but can accommodate any collection of sequences. So the first metagenome was loaded into CoGe from NCBI: | ||
[http://www.ncbi.nlm.nih.gov/nuccore/CABR00000000 Mine drainage metagenome, whole genome shotgun sequence] | |||
= | And can be seen in CoGe: http://genomevolution.org/CoGe/OrganismView.pl?oid=32988 | ||
== Assembling contig-level assembles to a reference genome using synteny == | |||
Feb. 18th 2011 | |||
[[SynMap]] has an option for generating a [[Syntenic path assembly]] with the click of a button. When complete, there is an option to print out your assembled genome. | |||
== CoGe 2011 [http://www.intl-pag.org/ Plant and Animal Genome] conference presentations available for download == | |||
Feb. 10th 2011 | |||
For a complete list of PAG sessions: http://www.intl-pag.org/19/19-workshops.html | |||
=== "CoGe: Comparative genomics made easy!" === | |||
[http://www.intl-pag.org/19/19-comp-genetics.html Comparative Genomics Workshop] | |||
Eric Lyons, iPlant Collaborative and University of Arizona, Tuscon AZ (ericlyons@e-mail.arizona.edu) | |||
PDF available at: http://genomevolution.org/CoGe/data/distrib/presentations/PAG-2011-CoGe-CompG.key.pdf | |||
<br> | |||
=== "10,000 Genomes at Your Fingertips" === | |||
[http://www.intl-pag.org/19/19-demos.html Computer Demonstrations] | |||
Eric Lyons, iPlant Collaborative and the University of Arizona, Tuscon AZ (ericlyons@email.arizona.edu) | |||
PDF available at: http://genomevolution.org/CoGe/data/distrib/presentations/PAG-2011-CoGe-ComputerDemp.key.pdf | |||
== Chocolate genome gene models added == | |||
Feb. 4th 2011 | |||
Thanks to [http://cocoagendb.cirad.fr/ CIRAD] for sharing their cacao gene models. These have been added to the Theobrama cacao genome in CoGe: http://genomevolution.org/CoGe/OrganismView.pl?dsgid=10997 . | |||
For an example of how these gene models may be used in whole genome comparisons, see this analysis between chocolate and peach: [[Chocolate-peach syntenic dotplots]]. It shows how the evolutionary distance between sytnenic gene pairs may be visualized to differentiate between [[Orthologous]] syntenic regions derived from the divergence of these lineages, and [[Out paralogous]] syntenic regions derived from their shared [[Paleohexaploidy]] ancestry. | |||
== Arabidopsis thaliana [http://arabidopsis.org TAIR] version 10 has been added! == | |||
Jan. 27th 2011 | |||
Version 10 of the Arabidopsis thaliana genome has been added to CoGe: http://genomevolution.org/CoGe/OrganismView.pl?dsgid=11022 | |||
Thanks to all the work by the folks at [http://arabidopsis.org TAIR] | |||
For a syntenic dotplot of version 9 versus version 10 of Arabidopsis thaliana (with the evolutionary distances of syntenic gene pairs calculated) see: http://genomevolution.org/r/2hiz | |||
== Chocolate genome added: from the [http://cocoagendb.cirad.fr/ International Cacao Genome Sequencing Consortium] == | |||
Jan. 26th 2011 | |||
The genome of Theobroma cacao has been published: http://www.nature.com/ng/journal/vaop/ncurrent/full/ng.736.html | |||
You can view this genome in CoGe at: http://genomevolution.org/CoGe/OrganismView.pl?dsgid=10997 | |||
To view some Syntenic dotplots of Cacao: [[Cacao syntenic dotplots]] | |||
Of note, this genome has not had any whole genome duplication events since the [[Paleohexaploidy]] event at the base of the eurosids. | |||
Both the 50x super masked and unmasked versions of the B73_refgen2 maize genome are now updated with the [http://ftp.maizesequence.org/current/README.txt new gene models] released by maizesequence.org over thanksgiving break. The new genome annotation consists of 110,028 genes, many with alternative transcripts, which can be broken down as followes: | == Version 2 of the Maize Genome, Now With Gene Models == | ||
*29,082 transposon related genes | |||
*17,615 putative pseudogenes | Both the 50x super masked and unmasked versions of the B73_refgen2 maize genome are now updated with the [http://ftp.maizesequence.org/current/README.txt new gene models] released by maizesequence.org over thanksgiving break. The new genome annotation consists of 110,028 genes, many with alternative transcripts, which can be broken down as followes: | ||
*29,082 transposon related genes | |||
*17,615 putative pseudogenes | |||
*63,276 "real" genes. Please note while these genes were annotated as "protein coding" in the current release, they include predicted microRNA genes. | *63,276 "real" genes. Please note while these genes were annotated as "protein coding" in the current release, they include predicted microRNA genes. | ||
==Maintenance Complete== | == Maintenance Complete == | ||
Sept. 16th 2010 | |||
Sept. 16th 2010 | |||
CoGe's servers have successfully be moved to a new rack space. | CoGe's servers have successfully be moved to a new rack space. Thanks to James, Bao, and Brent for making this happen. | ||
==Pending CoGe Maintenance== | == Pending CoGe Maintenance == | ||
Sept. 15th 2010 | |||
We have received word from the UC Data Center which houses CoGe that we need to move our servers to a new rack space. This should only take an hour or two. Our tentative schedule time for the move is: | |||
Sept 16th 2010 at 1pm (PCT) | |||
We apologize for any inconvenience this may cause any of CoGe's users. | |||
== [[SynMap]] update == | |||
Aug. 26th 2010 | |||
Organisms selected in [[SynMap]] have links in their taxonomic descriptions. If you click on a term in the taxonomic description, that term is automatically entered into the organism description search. All organisms with a matching taxonomic term will be displayed. This makes it faster to find organisms related to the one in which you are interested. | |||
<br> | |||
External searches under organism information: | == [[OrganismView]] update == | ||
*NCBI | |||
*Wikipedia | Aug. 26th 2010 | ||
[[OrganismView]] now has more links for finding information about an organism, and to internal CoGe tools. | |||
External searches under organism information: | |||
*NCBI | |||
*Wikipedia | |||
*Google | *Google | ||
Internal CoGe links: | Internal CoGe links: | ||
*[[CodeOn]]: | |||
*[[SynMap]]: | *[[CodeOn]]: automatically generates a table of amino acid usage as a function of the GC content of [[CDS]] sequences. | ||
*[[SynMap]]: (under Genome information) automatically loads SynMap with both genomes specified to the one selected. This makes is quick to start generating whole genome comparisons and [[Syntenic dotplots]]. | |||
== Home Page update == | |||
Aug. 26th 2010 | |||
CoGe's homepage menu "Latest Genomes" now has links to search for the organism name in | |||
*CoGe's [[OrganismView]] | |||
*CoGe's [[OrganismView]] | *NCBI | ||
*NCBI | *Wikipedia | ||
*Wikipedia | |||
*Google | *Google | ||
This makes it quicker to find information on an organism, specifically if you have no idea what it is. | This makes it quicker to find information on an organism, specifically if you have no idea what it is. Helpful considering that there are nearly 9,000 organisms in CoGe. | ||
== CoGeBlast update == | |||
Aug. 26th 2010 | |||
Aug. 26th 2010 | |||
[[CoGeBlast]] now has support for specifying blastn, tblastx, lastz, megablast, and discontinuous megablast when searching with nucleotide sequences. | [[CoGeBlast]] now has support for specifying blastn, tblastx, lastz, megablast, and discontinuous megablast when searching with nucleotide sequences. | ||
==10,000th genome loaded!== | == 10,000th genome loaded! == | ||
Aug. 4th 2010 | |||
= | [http://genomevolution.org/CoGe/OrganismView.pl?oid=32114 Brassica rapa] has been added to CoGe and represents the 10,000th genome loaded in CoGe. Its sequence was generated by the [http://www.genomics.cn/en/index.php BGI] located in China. This relative of Arabidopsis is a wonderful addition to sequenced plant genomes. Their lineage share a series of whole genome duplication events (commonly known as alpha, beta, and gamma -- the latter happening prior to the radiation of the eudicots). Since their divergence, [[Brassica rapa triploidy|Brassica rapa has had a triploidy]] while [[B.rapa versus A. thaliana|Arabidopsis has had none]]. | ||
== Genome update from NCBI == | |||
June 28th 2010 | |||
June | |||
A new update of genomes from NCBI has finished. This includes genomes from all domains of life. CoGe now has genomic sequence from 8,872 organisms comprising 9,999 genomes. There is also a new option on the homepage to list the most recently added genomes. | |||
== SIP 2010 workshop syllabus == | |||
June 23rd 2010 | |||
June | |||
The | The syllabus for a day-long workshop on how to use CoGe for [http://www.sip2010.org/ the Society for Invertebrate Pathology's conference (SIP 2010)] is now available. This workshop focuses on: | ||
#Getting an overview of how CoGe is designed for allowing scientists to create their own open-ended analyses | |||
#Learning what the various tools in CoGe do and they to use them | |||
#Working through specific sets of example problems focused on analyzing two groups of organisms important for invertebrate pathology: baculoviruses and Bacillus thuringiensis | |||
This new version of CoGe has: | The workshop's syllabus is available: [[SIP2010]] | ||
#Update UI | |||
#Various feature extensions on existing tools | == CoGe's update progress == | ||
#Updated algorithms (new blast API with support for the megablast families, LastZ) | |||
#New database additions | June 18th 2010 | ||
#Update of core modules for database API | |||
The switch to the new server went as smoothly as I could have hoped. | |||
Besides from new hardware (which should greatly accelerate many of CoGe's analyses and improve system stability), this installation welcomes a new version of CoGe too! | |||
This new version of CoGe has: | |||
#Update UI | |||
#Various feature extensions on existing tools | |||
#Updated algorithms (new blast API with support for the megablast families, LastZ) | |||
#New database additions | |||
#Update of core modules for database API | |||
#New configuration files that will help deployment of CoGe to new sites | #New configuration files that will help deployment of CoGe to new sites | ||
Please contact [mailto:elyons@berkeley.edu Eric Lyons] if you find any bugs! | Please contact [mailto:elyons@berkeley.edu Eric Lyons] if you find any bugs! | ||
== Today is the day == | |||
June 17th 2010 | |||
Going to through the switch today. Expect some downtime with CoGe and some support systems being temporarily off line. | |||
== New CoGe Server Update == | |||
June 10th 2010 | |||
June | |||
It appears that most of the software updates and migration to the new server are working. We have deployed the new server to the UC Data Center, but due to some complications with rack-space, IP address allocation, sub-nets, firewalls, etc., things may be in flux for a while. We've had to take our development server (aka toxic) off line and put the new server on its IP address till those things get sorted out. In the meanwhile, we will plan on making the switch to production on the new server soon (hopefully next week). When this happens, expect CoGe to be offline for a couple of hours, but we will do our best to keep downtime to a minimum. | |||
==New CoGe Server | == New CoGe Server is being readied! == | ||
June 2nd 2010 | |||
We have our new server for CoGe! Its deployment will not only include new performance improvements due to more computing power, but all several changes and additions to CoGe: | |||
#new user interface | |||
#new user interface | #new algorithm options | ||
#new algorithm options | |||
#new structure of the underlying code-base to make it is easier to redeploy (in anticipation of eventually getting the code-base released to those interested) | #new structure of the underlying code-base to make it is easier to redeploy (in anticipation of eventually getting the code-base released to those interested) | ||
We are planning on moving the new server to the UC data center this Fri. | We are planning on moving the new server to the UC data center this Fri. After some more testing and bug hunting, we will switch our current production server's IP address to this machine. There is a high chance that there will be some downtime for CoGe during this switch and we will post announcements as to when this change will happen! In the meanwhile, if anyone is interested in testing new CoGe, please e-mail [mailto:elyons@berkeley.edu Eric Lyons]. | ||
== SGRP: (Sanger Institute) yeast genomes added to CoGe == | |||
May 18th 2010 | |||
75 Yeast genomes from [http://www.sanger.ac.uk/research/projects/genomeinformatics/browser.html SGRP (Saccharomyces Genome Resequencing Project)] have been added to CoGe. For a complete list of Organisms, please see [[SGRP: Sanger Institute Yeast Genomes]]. | |||
== CoGe post on The OpenHelix == | |||
May 5th 2010 | |||
Eric Lyons wrote a piece about CoGe for [http://blog.openhelix.eu/?p=4276 The OpenHelix Blog] | |||
== Version 2 of [[Sequenced plant genomes#Maize.2FCorn|Maize B73]] genome added to CoGe == | |||
May 3rd 2010 | |||
= | <font color="red">This release does not yet have annotations (yet)</font>! | ||
You can view the genome in CoGe at: http://genomevolution.org/CoGe/OrganismView.pl?dsgid=9106 | |||
This sequences was obtained from: http://www2.genome.arizona.edu/genomes/maize | |||
And can read about differences in the assembly between versions 1 and 2: [[Maize v1 v2|here.]] | |||
==Version 2 of [[ | == Version 2 of [[Sequenced plant genomes#Grape|Vitis vinifera (grapevine)]] genome added to CoGe == | ||
Apr. 10th 2010 | |||
You can view the genome in CoGe at: http://genomevolution.org/CoGe/OrganismView.pl?dsgid= | You can view the genome in CoGe at: http://genomevolution.org/CoGe/OrganismView.pl?dsgid=9048 | ||
Version 2 with 12x coverage was obtained from [http://www.genoscope.cns.fr/externe/Download/Projets/Projet_ML/data/12X/ Genoscope]. | |||
There are some changes to the assembly with new contig orders and additional sequence added to the pseudomolecules which can been seen [[Vitis vinifera version 2 versus version 1|here]]. | |||
== | == New NCBI Genome Update. CoGe surpasses 8,900 genomes from 8,200 organisms == | ||
Apr. 9th 2010 | |||
Finished an update from NCBI. However, this is not a complete listing of all genomes available at NCBI due to some API problems getting some genomes. You can read about this problem below. | |||
== Version 3 of [[Sequenced plant genomes#Medicago|Medicago truncatula]] added to CoGe == | |||
Apr. 9th 2010 | |||
Apr. 9th 2010 | |||
You can view the genome in CoGe at: http://genomevolution.org/CoGe/OrganismView.pl?dsgid=8976 | |||
[[Syntenic dotplot medicago truncatula version 3 versus version 2|Syntenic comparison of version 3 to version 2]] shows extensive changes in the primary sequence. Some chromosomes have had their sequence substantially updated. | |||
== [[Sequenced plant genomes#Peach|Prunus persica (peach tree)]] added to CoGe == | |||
Apr. 9th 2010 | |||
You can view its genome in CoGe at: http://www.genomevolution.org/CoGe/OrganismView.pl?oid=30980 | |||
Its genome was produced by [http://www.peachgenome.org the International Peach Genome Initiative] and its sequence was obtained from [ftp://ftp.jgi-psf.org/pub/JGI_data/phytozome/v5.0/Ppersica/ phytozome]. This genome is currently [[Sequenced plant genomes#Peach|unpublished]] and therefore under the publication restrictions of the [[Fort Lauderdale Convention]]. | |||
Peach is a eudicot in the Rosaceae family. | |||
== Automatic NCBI Genome Loader Update == | |||
Apr. 8th 2010 | |||
Apr. 8th 2010 | |||
The automatic NCBI genome loader is running today. | The automatic NCBI genome loader is running today. It has been a while since I last ran it after running into an API problem with NCBI's eutils tools three months ago. The issue is still unresolved and even after checking in every two weeks for a status update, I have yet to receive any word as to when the bug will be fixed. For those interested, here is my bug report sent at the end of January: | ||
Issue (http://jira.be-md.ncbi.nlm.nih.gov/browse/HD-1843): | Issue (http://jira.be-md.ncbi.nlm.nih.gov/browse/HD-1843): | ||
Key: HD-1843 | |||
Summary: Unable to get some genomes using eutils | |||
Type: Task | |||
Status: In Progress | |||
Priority: Normal | |||
Assignee: Matten, Wayne | |||
Reporter: Nobody | |||
Description: | Description: | ||
| Line 925: | Line 1,032: | ||
accessions CPXXXXXX) are have a genomeprj id but no associated genome id. For example, genomeprj=30031. | accessions CPXXXXXX) are have a genomeprj id but no associated genome id. For example, genomeprj=30031. | ||
It is listed in this list: http://eutils.ncbi.nlm.nih.gov/entrez/eutils/esearch.fcgi?db=genomeprj&term=all%5Bfilter%5D&retmax=999999 | It is listed in this list: http://eutils.ncbi.nlm.nih.gov/entrez/eutils/esearch.fcgi?db=genomeprj&term=all%5Bfilter%5D&retmax=999999 | ||
But has no genome id: http://eutils.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?db=genome&dbfrom=genomeprj&id=30031 | But has no genome id: http://eutils.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?db=genome&dbfrom=genomeprj&id=30031 | ||
However, it does have an entry in genbank: | However, it does have an entry in genbank: | ||
http://www.ncbi.nlm.nih.gov/nuccore/CP001637.1?ordinalpos=3&itool=EntrezSystem2.PEntrez.Sequence.Sequence_ResultsPanel.Sequence_RVDocSum | http://www.ncbi.nlm.nih.gov/nuccore/CP001637.1?ordinalpos=3&itool=EntrezSystem2.PEntrez.Sequence.Sequence_ResultsPanel.Sequence_RVDocSum | ||
I am probably missing something obvious. Can you help me figure out how to get a list of all the genomes at NCBI? I am using | I am probably missing something obvious. Can you help me figure out how to get a list of all the genomes at NCBI? I am using | ||
| Line 941: | Line 1,048: | ||
-Eric Lyons | -Eric Lyons | ||
If anyone has any solutions to this problem, please contact me. | If anyone has any solutions to this problem, please contact me. | ||
==Major bug fix in [[SynMap]]== | == Major bug fix in [[SynMap]] == | ||
Mar. 26th 2010 | |||
While testing the prior bug fix, I discovered that [[SynMap]] wasn't working on genomic sequence comparisons (as opposed to CDS sequence comparisons). This was due to the new analytical pipeline's data processing requiring unique names for each blast hit. Otherwise, multiple hits to the same sequence name would get removed as a [[Local duplicate]]. As all hits to a genomic sequence were named according to the chromosome, all such hits were flagged as [[Local duplicates]] and removed from the analysis. | |||
As always, if you find a problem in CoGe, feel free to email [mailto:elyons@berkeley.edu Eric Lyons] and let him know what you've found. There are now too many options and buttons to click in CoGe for me to test with each update. | |||
== Minor bug fix in [[SynMap]] == | |||
Mar. 25th 2010 | |||
Mar. | |||
With SynMap's new analytical pipeline, there are still some bugs to be worked through. Hopefully got one today in the script that converted blast input files to [http://genome.ucsc.edu/FAQ/FAQformat.html#format1 bed format], which is required for the program to find local duplicates in the compared genomes. These local duplicates are removed from the algorithm for finding collinear series of putative homologous genes used to infer syntenic regions. Also, these local duplicate files are displayed in the download section of the results in case they are wanted for other analyses. | |||
== | == Hosting local tiny URL encoding == | ||
James Schnable has created a page detailing all of the sequenced plant genomes including: | Mar. 24th 2010 | ||
*overview of their genomic content | |||
*publications | Replaced using tinyurl.com for a local installation of a URL hashing and redirecting service. Makes generating these faster and allows for customized names. Note: the tinyurls will still work. | ||
*status of completion | |||
== [[Sequenced plant genomes]] == | |||
Mar. 13th 2010 | |||
James Schnable has created a page detailing all of the sequenced plant genomes including: | |||
*overview of their genomic content | |||
*publications | |||
*status of completion | |||
*interesting factoids (e.g. The average US American eats 25lbs of bananas a year.) | *interesting factoids (e.g. The average US American eats 25lbs of bananas a year.) | ||
Read about them here: [[Sequenced plant genomes]] | Read about them here: [[Sequenced plant genomes]] | ||
== The JGI's [[Sequenced plant genomes#Cassava|Manihot esculenta (cassava)]] genome has been added == | |||
Mar. 13th 2010 | |||
This genome from the [http://www.jgi.doe.gov/ JGI] brings CoGe up-to-date with [http://www.phytozome.net phytozome] v5.0. | |||
You can access cassava in CoGe [http://synteny.cnr.berkeley.edu/CoGe/OrganismView.pl?oid=30762 here], and get more information from [http://www.phytozome.net/cassava.php phytozome]. | |||
== The JGI's [[Sequenced plant genomes#Cassava|Cucumis sativus (cucumber)]] genome has been added == | |||
Mar. 12th 2010 | |||
Mar. 12th 2010 | |||
You can access it in CoGe [http://synteny.cnr.berkeley.edu/CoGe/OrganismView.pl?dsgid=8164 here]. | You can access it in CoGe [http://synteny.cnr.berkeley.edu/CoGe/OrganismView.pl?dsgid=8164 here]. Or get more information about it from [http://www.phytozome.net/cucumber.php phytozome]. This is apparently a distinct sequence from the one [http://dx.doi.org/10.1038/ng.475published in Nature Genetics last November.] That sequence was from "'Chinese long' inbred line 9930" this version comes from the inbred Gy14. More details [[Sequenced plant genomes#Cucumber|here]] | ||
==[[SynMap]] updated== | == [[SynMap]] updated == | ||
After a month of work, [[SynMap]] has undergone several significant changes, incorporating new algorithms written by Haibao Tang and Brent Pedersen: | Mar. 12th 2010 | ||
*new merging function for overlapping and neighboring diagonals (program: | |||
*new method for detected tandem gene duplicates | After a month of work, [[SynMap]] has undergone several significant changes, incorporating new algorithms written by Haibao Tang and Brent Pedersen: | ||
*new merging function for overlapping and neighboring diagonals (program: quota alignment) | |||
*new method for detected tandem gene duplicates | |||
*better reporting of all intermediate files used in the analysis, including tandem duplicates | *better reporting of all intermediate files used in the analysis, including tandem duplicates | ||
These changes have also hoped to increase the stability of SynMap, which due to its long pipeline, has been known to crash for some genomes and/or specific parameter configurations. | These changes have also hoped to increase the stability of SynMap, which due to its long pipeline, has been known to crash for some genomes and/or specific parameter configurations. Please let [mailto:elyons@berkeley.edu Eric Lyons] know if you have any problems with an analysis. Please send along the names of the organisms/genomes compared and a copy of the log file produced by each SynMap run (if possible). | ||
== Persistent GEvo bug fixed == | |||
Mar. 11th 2010 | |||
A long-stranding, but intermittent and annoying bug in [[GEvo]] has finally fixed. This (hopefully) solves the problem where once in a while, [[GEvo]] will return blank results to its interactive viewer, [[Gobe]]. The crux of the bug, and why it was intermittent (and hence difficult to reproduce and trouble-shoot), was a race condition between asynchronous client javascript code and server perl code. Perl was responsible for generating a random session id for the analysis, but it occasionally failed to return that id to the client code before the analysis was sent back to the server for processing. When this happened, the processing analysis received a default id and multiple analyses could be merged if the default id had been used within that past 24 hours (the length of time an analysis stays on the server before being deleted). When [[Gobe]] tried to process the results, the stored data and what was specified for initialization did not match, thus causing gobe to fail and return blank results. The solution: have javascript generate the analysis session id so there is no chance of a delay before the analysis is sent to the server for processing. | |||
However, if anyone does come across this bug again (or any others), please let me know: [mailto:elyons@berkeley.edu Eric Lyons] | |||
== Rice Version 6.1 loaded == | |||
Mar. 10th 2010 | |||
= | You can view it in [http://toxic.berkeley.edu/CoGe/OrganismView.pl?dsgid=8163 GenomeView]. This was retrieved from [http://rice.plantbiology.msu.edu/index.shtml MSU's Rice Genome Annotation Project]. | ||
== The classic set of Maize Genes == | |||
Mar. 9th 2010 | |||
[[Classical Maize Genes|The classical maize gene list]] | |||
[[User:Jschnable|James Schnable]] manually evaluate ~460 classic maize genes available from [http://maizegdb.org MaizeGDB] and [http://www.ncbi.nlm.nih.gov/ NCBI], determined their genomic positions in the maize genome, and found their [[Syntenic]] regions within maize (from its most recent [[Whole genome duplication event]]), sorghum, rice, and brachypodium. [[Classical Maize Genes|This list]] contains links to compare these syntenic regions using [[GEvo]]. | |||
== | == New plant genomes in CoGe == | ||
Feb. 10th 2010 | |||
'''Mimulus guttatus''' (monkey flower): http://synteny.cnr.berkeley.edu/CoGe/OrganismView.pl?oid=30760 Mimulus is an outgroup to the rosids (in the sister group, the asterids) | |||
'''Populus trichocarpa''' (Poplar; cotton wood): http://synteny.cnr.berkeley.edu/CoGe/OrganismView.pl?oid=324 Version 2 of poplar! | |||
Both are from the [http://www.jgi.doe.gov/ JGI]. | |||
== MaizeGDB links to [[GenomeView]] == | |||
Feb. 8th 2010 [http://maizegdb.org MaizeGDB] is now linking to CoGe's [[GenomeView]] so maize researchers can find maize-sorghum [[Syntenic gene sets]] and quickly perform syntenic analyses using [[GEvo]]. For an example view from MaizeGDB's genome broswer: | |||
http://gbrowse.maizegdb.org/cgi-bin/gbrowse/maize/?name=chr1:1000000..1200000 | |||
For instructions on how to perform this workflow: [[MaizeGDB and CoGe]] | |||
For | For more information on maize-sorghum syntenic analyses: [[Maize Sorghum Syntenic dotplot|Maize-Sorghum genome analyses]] | ||
For | For a quick video walk through of the new connections: [[Tutorials#MaizeGDB_and_CoGe.27s_Maize-Sorghum_Orthologies|MaizeGDB_and_CoGe.27s_Maize-Sorghum_Orthologies]] | ||
== Syntelog visualization in [[GenomeView]] == | |||
Feb. 5th 2010 [[GenomeView]] has been updated to auto-detect [[Genomic features]] with annotations that are links to [[GEvo]]. These links provide an analysis of a [[Genomic feature]] (e.g. gene) to previously identified [[Syntologous]] sets of features. Currently, this has been implemented using syntelogs from [[Maize Sorghum Syntenic dotplot|maize and sorghum]], but with the code in place, we will expand annotations for genomic features from other organisms for which we generated syntologous gene sets. For an example of this visualization in [[GenomeView]] please see: [http://synteny.cnr.berkeley.edu/CoGe/GenomeView.pl?fid=19472659_1&dsid=34580&chr=&x=&dsgid=93;z=5 | this GenomeView of sorghum]. Also, for an expanded list of glyphs used in [[GenomeView]] please refer [[GenomeView examples|to these examples]]. | |||
Feb. 5th 2010 | |||
[[GenomeView]] has been updated to auto-detect [[ | |||
==Easy exporting and downloading of genomes== | == Easy exporting and downloading of genomes == | ||
Jan. 16th 2010 [[OrganismView]] has new options for easily downloading the sequences of a genome in fasta format and retrieving all of its annotations in an GFF file. To access, just search for an organism and genome of interest, and look for the links under "Genome Information". | |||
We've linked to [http://www.phylogeny.fr phylogeny.fr] for quick and easy phylogenentic tree reconstruction. | == [[FastaView]] is linked to [http://www.phylogeny.fr phylogeny.fr] for one-click phylogenetics == | ||
#multiple sequence alignment (MUSCLE) | |||
#maximum likelihood phylogenetic tree reconstruction (PhyML) | Jan. 10th. 2010 | ||
We've linked to [http://www.phylogeny.fr phylogeny.fr] for quick and easy phylogenentic tree reconstruction. Now, you can build a list of fasta sequences and display them in [[FastaView]], select protein or DNA sequences, edit them if necessary (e.g. add or remove sequences manually), and press a button to send them off to phylogeny.fr for: | |||
#multiple sequence alignment (MUSCLE) | |||
#maximum likelihood phylogenetic tree reconstruction (PhyML) | |||
#tree visualization (TreeDyn) | #tree visualization (TreeDyn) | ||
For an example, use [http://synteny.cnr.berkeley.edu/CoGe/FastaView.pl?featid=2575204_1&featid=2575202_1&featid=37282168_1&featid=37282170_1&featid=6731432_1&featid=6731434_1&featid=6921617_1&featid=6923982_1&featid=6923986_1&featid=6934003_1&featid=6934001_1&featid=6936384_1&featid=6936382_1;prot=1 this link to FastaView] and press the button "phylogeny.fr" at the bottom of the screen. | For an example, use [http://synteny.cnr.berkeley.edu/CoGe/FastaView.pl?featid=2575204_1&featid=2575202_1&featid=37282168_1&featid=37282170_1&featid=6731432_1&featid=6731434_1&featid=6921617_1&featid=6923982_1&featid=6923986_1&featid=6934003_1&featid=6934001_1&featid=6936384_1&featid=6936382_1;prot=1 this link to FastaView] and press the button "phylogeny.fr" at the bottom of the screen. | ||
Special thanks to Haibao Tang for pointing out this incredible web resource! | |||
== Haibao Tang joins the Freeling lab == | |||
Jan. 4th 2010 | |||
Haibao Tang, an expert in plant comparative genomics and genome evolution, as well as a great python programmer, has joined the Freeling lab. His input and contributions will be most valued! | |||
== New [[Tutorials]] added == | |||
Jan. 4th 2010 | |||
New [[Tutorials]] have been added: | |||
*[[Tutorials#How_to_find_syntenic_regions_between_genomes.3F|How to find syntenic regions between genomes]] | |||
*[[Tutorials#Finding_Inversions|How to find inversions]] | |||
*[[Tutorials#Finding_rarely_and_frequently_used_codons_in_a_genome|How to find rarely and frequently used codons in a genome]] | |||
*[[Tutorials#Generating_an_amino_acid_usage_table_for_an_organism|How to generate an amino acid usage table for a genome]] | |||
*[[Tutorials#Whole_Genome_Comparison_and_Analysis_using_SynMap_and_GEvo|Using synonymous mutation rates in SynMap to rapidly identify different whole genome evolutionary events]] | |||
*[[Tutorials#How_to_extract_all_the_gene_sequences_from_a_genomic_region_for_export_from_CoGe|How to extract all gene sequences from a genomic region]] | |||
*[[Tutorials#Identifying_putative_horizontal_gene_transfer_events|How to identify putative horizontal gene transfer events]] | |||
== Linked to ProSite for protein domain searching == | |||
Dec. 24th 2009 | |||
== | FastaView is now linked to [http://www.expasy.ch/prosite/ ProSite] when viewing a protein sequence for protein domain searching. See [http://synteny.cnr.berkeley.edu/CoGe/FastaView.pl?featid=9346534&gstid=1;prot=1 this FastaView example] and click on the link at the bottom of the page. | ||
== Improved implementation of DAGChainer in [[SynMap]] == | |||
Dec. 15th 2009 | |||
Thanks again to Brent Pedersen for some fantastic programming. He discovered that DAGChainer's C++ code's makefile did not include the -O3 optimization, rewrote the input/output methods of the compiled binary to read from STDIN instead of a file, and rewrote the perl front-end in python. Together, these changes increase CoGe's DAGChainer implementation in SynMap between 2-4 fold. | |||
You can download his code at: svn co http://bpbio.googlecode.com/svn/trunk/scripts/dagchainer | |||
== CoGe Workshop being taught at SIP 2010 == | |||
Nov. 30th 2009 | |||
Genomics: What every invertebrate pathologist needs to know. http://www.sip2010.org/index.php/Bioinformatics-Workshop.html | |||
== CoGe on OpenHelix and James and the Giant Corn == | |||
Nov. 18th 2009 | |||
Nov. | |||
Phillipe Lamesch from TAIR passed along a link to [http://blog.openhelix.com/?p=2913 openhelix.com] highlighting CoGe's tool GEvo. They put together a nice [http://www.openhelix.com/downloads/jing/gevo.swf video] showing GEvo. They, in turn, found this on a posting at the blog of [http://www.jamesandthegiantcorn.com/2009/11/07/obama-on-nsf-fellowships/ James and the Giant Corn] who had used GEvo for a grant proposal. | |||
http://www. | |||
== | == Maize Pseudomolecule Assembly with Gene Models Released == | ||
Oct. 20th 2009 | |||
Thanks to maizesequence.org [http://ftp.maizesequence.org/release-4a.53/ for providing the sequence and annotations.] The current pseudomolecule assembly of maize has been loaded into CoGe. | |||
*[http://synteny.cnr.berkeley.edu/CoGe/OrganismView.pl?dsgid=8054 Link to OrganismView for complete set of gene models.] | |||
*[http://synteny.cnr.berkeley.edu/CoGe/OrganismView.pl?dsgid=8054 Link to OrganismView for complete set of gene models.] | *[http://synteny.cnr.berkeley.edu/CoGe/OrganismView.pl?dsgid=8053 Link to OrganismView for filtered gene model set.] | ||
*[http://synteny.cnr.berkeley.edu/CoGe/OrganismView.pl?dsgid=8053 Link to OrganismView for filtered gene model set.] | *[http://synteny.cnr.berkeley.edu/CoGe/SynMap.pl?dsgid1=8053;dsgid2=93;D=20;g=10;A=5;w=0;b=1;ft1=1;ft2=1;dt=geneorder;ks=1;autogo=1 Maize-Sorghum syntenic dotplot with syntologs colored by synonymous rate change.] | ||
*[http://synteny.cnr.berkeley.edu/CoGe/SynMap.pl?dsgid1=8053;dsgid2=93;D=20;g=10;A=5;w=0;b=1;ft1=1;ft2=1;dt=geneorder;ks=1;autogo=1 Maize-Sorghum syntenic dotplot with syntologs colored by synonymous rate change.] | |||
*[http://synteny.cnr.berkeley.edu/CoGe/SynMap.pl?dsgid1=8053;dsgid2=8053;D=20;g=10;A=5;w=0;b=1;ft1=1;ft2=1;dt=geneorder;ks=1;autogo=1 Maize-Maize syntenic dotplot with syntologs colored by synoonymous rate change.] | *[http://synteny.cnr.berkeley.edu/CoGe/SynMap.pl?dsgid1=8053;dsgid2=8053;D=20;g=10;A=5;w=0;b=1;ft1=1;ft2=1;dt=geneorder;ks=1;autogo=1 Maize-Maize syntenic dotplot with syntologs colored by synoonymous rate change.] | ||
==CoGe surpasses 7000 organisms in its database!== | == CoGe surpasses 7000 organisms in its database! == | ||
More fun for everyone! | |||
More fun for everyone! | |||
== NCBI Genome Loader Updated == | |||
CoGe's automated NCBI genome loader has been updated and is once again checking NCBI regularly for new and updated genomes. You can get a snapshot of the number or organisms and genomic sequence in CoGe by checking its [http://synteny.cnr.berkeley.edu/CoGe homepage], search for your genome of interest using [[OrganismView]]. | |||
== CoGe is linked to [http://target.iplantcollaborative.org/ TARGeT: Tree Analysis of Related Genes and Transposons] == | |||
You can send a set of fasta sequence generated by [[FastaView]] directly to TARGeT. | |||
== | == New version of Gobe release! == | ||
Read general announcement [[Gobe]]. Major feature: transparent wedges are drawn to connect regions of sequence similarity. | |||
Read general announcement [[Gobe]]. | |||
== Version 3 of CoGe is released! == | == Version 3 of CoGe is released! == | ||
Read general announcement [[CoGe version 3]]. | Read general announcement [[CoGe version 3]]. | ||
Revision as of 21:10, 15 October 2012
CoGeBlast Update
Oct. 15th, 2012
The CoGeBlast user interface was revamped for a cleaner appearance and simpler use. The functionality remains unchanged, except the addition of a button to import target genomes from existing lists.
CoGe "Data Tab"
Oct. 5th, 2012
With the roll-out of CoGe v5 comes the ability for users to more easily organize and share their data of interest. Most of these features are found in the "Data" tab in the CoGe menu located in the upper right part of the screen:
- User Profile: Shows what information CoGe's stores about you (user name, real name, email address) and a list of your groups
- User Groups: Groups of users to which you have access. These groups are used to share lists of data
- Data Lists: Lists of data (genomes, features, experiments) to which you have access. May be private or public data. It is through these lists and User Groups that allows you to share private data with collaborators
- History: CoGe has always generated tiny links for your analysis and views of data. These are now stored for you so you may more easily find a previously run analysis.
CoGe v5 Deployment Process
Sept 24th, 2012
- 9am: We shut down the website at 9am and started the final backup and freezing of existing data and analyses.
- 10am: Database is being replicated to the iPlant Data Store and copied to a backup server for processing and conversion to new database scheme.
- 11am: All web-code and libraries were backed up and new code deployed
- 12pm: updating database
- 1pm: copying database back to iRODS
- 2pm: copying database to coge server
- 3pm: reconfiguring the system
- 3:30pm: turn on web server
- 3:31pm: Nothing works
- 3:32pm: Start debugging
- 3:34pm: Get things working -- CoGe starts!
- 4:30pm: Most major problems found and corrected
CoGe v5 Deployment
Sept 21st, 2012
CoGe v5 is planned for deployment on Sept. 24th. This new version of CoGe represents a massive revamping and extension of the user-data management system.
Key features include:
- Limited support for experimental data
- This new system is funded (in part) by a grant from the Gordon and Betty Moore Foundation to add visualization support for epigenetics data for Arabidopsis. This is known as the EPIC-CoGe project: http://www.iplantcollaborative.org/learn/news/2012/05/24/iplant-ci-leveraged-development-epic-coge-browser
- Ability to make lists and collections of data
- Lists of experiments
- Lists of genomes
- Lists of features
- Lists of lists
- Enhancements for managing and sharing private data in CoGe
- Logging user history so it is easier to find old analyses
A key part to the migration to the new version of CoGe is preserving current private data in the system and assigning them to the appropriate owner. (We have done some major changes to the underlying metadata storage database for CoGe). Please let us know if you have lost access to your data and we will get that corrected right away.
New features will be added that further integrate user specified lists into various tools in CoGe. E.g. auto-selecting a list of genomes for use in CoGeBlast instead of manually searching for all the genomes.
Many thanks to CoGe Developer Matt Bomhoff for all the work on this new version.
Please post any comments, suggestion, questions to CoGe's Forums (hosted by iPlant): https://forums.iplantcollaborative.org/viewforum.php?f=10
Phaseolus vulgaris (common bean) v1 added to CoGe
Aug 23, 2012
Released from JGI/Phytozome, it is v1 of the common bean: http://genomevolution.org/CoGe/OrganismView.pl?oid=36223
Syntenic dotplots between it and soybean (Glycine max), Phaseolus vulgaris v. Glycine max, clearly show that Phaseolus lacks the most recent Whole genome duplication in the Glycine lineage.
CoGe Paper published: Unleashing the genome of Brassica rapa
July 31th, 2012
This Open Access paper provides a set of examples of how to analyze and compare the genome of Brassica rapa. Very useful for people wanting to learn how to use CoGe or how to maximize their use of the genome of Brassica rapa:
Open Access article in Frontiers of Plant Genetics and Genomics: http://www.frontiersin.org/plant_genetics_and_genomics/10.3389/fpls.2012.00172/abstract
Also located in CoGe Tutorials sections.
Banana genome published
July 12th, 2012
The banana genome was published today in Nature: http://www.nature.com/nature/journal/vaop/ncurrent/full/nature11241.html
CoGe was used in some of the analyses (in supplementary figures), and the genome is now publicly available: http://genomevolution.org/coge/OrganismView.pl?oid=38351
Banana represents the first non-grass monocot genome to be sequenced and sheds light in the evolutionary history of the lineage as a whole.
My opinion is that the timing, placement, and make-up of the early monocot duplication events are still an open question. Some work points to an additional polyploidy event in the Poales lineage (See: Tang. et al. Angiosperm genome comparisons reveal early polyploidy in the monocot lineage. PNAS 2010; http://www.pnas.org/content/107/1/472.full). Banana, with its own series of independent series of whole genome duplications, is not the best suited for unravelling this earlier events, but these events open many avenues for additional research into the evolution and architecture of plant genomes. It will be exciting to see what similarities and differences exist between the monocots and the dicots.
Additional news pieces on banana:
 |
 |
Tomato genome published; the solanum hexaploidy investigated with CoGe
May 31st, 2012
The tomato genome was published in Nature earlier this week: http://www.nature.com/nature/journal/v485/n7400/
However, the current version of the tomato genome has been in CoGe for the past year (thanks to an early release of the data from the tomato genome consortium).
I've received a couple of emails inquiring about the Solanum specific hexaploidy, and this has been investigated with Haibao Tang. Overall, these analyses support that the majority of the genome is derived from a tetraploidy, but there is evidence of some regions being triplicated (perhaps through a hexaploidy).
These analyses are available: Tomato genome
Please send us your thoughts or post them on the CoGe Forum:
May 29th, 2012
If you want to load a private genome into CoGe, you need to send that genome to the CoGe team. This method makes it very easy for us to download your genome quickly!
Dedicate CoGe Forum hosted at iPlant (part of the powered by iPlant program)
May 25th, 2012
iPlant has set up a dedicated forum for CoGe: https://forums.iplantcollaborative.org/viewforum.php?f=10
Please post any CoGe questions you have to here.
EPIC-CoGe Browser
May 24th 2012
News article about this project at iPlant: http://www.iplantcollaborative.org/learn/news/2012/05/24/iplant-ci-leveraged-development-epic-coge-browser
Overview of the Epic-CoGe Browser prototype system:
Try it: http://genomevolution.org/CoGe/GenomeView.pl?z=6&x=20000&dsgid=7043&chr=1
WARNING: performance is a known issue! Some tiles in the browser may take a while to render (but are then cached).
2000 new genomes in CoGe
Apr. 30th 2012
The NCBI genome loader program was updated and run over the weekend. This resulting in about 2000 new genomes being loaded into CoGe.
Unscheduled CoGe downtime
Apr. 25th 2012
CoGe was down/offline yesterday for two reasons:
- One of iPlant's VMs was compromised and UITS (UA's IT group) shut off one of iPlant's subnets, which CoGe happens to use. This was due to a VM administered by a group collaborating with iPlant and not due to iPlant
- Since CoGe was offline, when it came up, we decided to keep it offline for a while longer in order to updated the apache web server. After apache was updated, CoGe was brought online. Unfortunately, UITS detected a security vulnerability in the SSL implementation in the new update and shut CoGe off. This last part happened at the end of the day and we weren't able to coordinate with UITS to push a fix until this morning.
While the CoGe team tries to keep as much uptime as possible, this type of downtime does happen once and a while. Our apologies to everyone whose work was interrupted or delayed due to this.
The algorithm, Last, added to SynMap
Mar. 28th 2012
Last (http://last.cbrc.jp/) has been added as a comparison algorithm in SynMap. Its performance is phenomenal! This is still under testing, so please let us know if you have any problems with it. Also, special thanks to Haibao Tang for writing the parallelized adapter for Last that is used by SynMap. Without this program, the integration would not have happened as quickly, easily, or smoothly.
CoGe used to decode the secret message in JCVI Synthetic Genome
Mar. 24th 2012
I heard that there was a secret message in the JCVI synthetic genome: Mycoplasma mycoides JCVI-syn1.0. Using CoGe, the DNA containing the secret messages was identified and decoded. Here is the walk-through of how this was done: Mycoplasma mycoides JCVI-syn1.0 Decoded.
- WARNING: contains spoilers!
- Note: this puzzle is nearly 2 years old.
For those interested in doing the puzzle, this article has a good summary of the challenge:
- Article from Sigularity Hub on the secret message: http://singularityhub.com/2010/05/24/venters-newest-synthetic-bacteria-has-secret-messages-coded-in-its-dna/
And you will probably need the original article (and the Supplementary Data):
- Original Science Article on the genome: http://www.sciencemag.org/content/329/5987/52.abstract
CoGe Forums
Mar. 2nd 2012
iPlant has a forums site available: http://forums.iplantcollaborative.org
CoGe, being part of the "Powered by iPlant" program, has a section on there for users to post questions about how to do various tasks, about CoGe in general, and provide suggestions. I'll be posting questions that are emailed to me there, but this will hopefully be a good place for people to ask questions, find answers, and help one another.
Powered by iPlant Forum: https://forums.iplantcollaborative.org/viewforum.php?f=8
The CoGe Forum: https://forums.iplantcollaborative.org/viewforum.php?f=10
BlastN Bug
Feb. 11th 2012
Mike Freeling from UC Berkeley has found an interesting bug in BlastN where a relatively large blast hit (HSP) appears/disappears depending on the amount of sequence compared between Arabidopsis and Brassica. James Schnable from UC Berkeley further characterized this by identifying a comparison that differs in 1 nucleotide (over ~750) that causes this effect. You can see images of this blast error, characterization of the blast, an breakdown of parameters used here: GEvo Blastn Bug
CoGe Server Migration
Feb. 4th 2012
CoGe's entire system has been migrated to the new server hosted by the [iplantcollaborative.org iPlant Collaborative]. This include
- A new version of CoGe (v4) that includes:
- Migration of CoGePedia to new server
- Migration of CoGe's tiny-url service (used to construct URLs that can be used to regenerated analysis -- mainly by GEvo and SynMap)
- Update to DNS for:
Please contact us if you come across any problems!
Exciting new plant genomes in CoGe
Feb. 3rd 2012
Update on genomes available from Phytozome.
The genomes of
- common bean (a crucial staple food of grad students everywhere)
- capsella (the close relative of arabidopsis, not the song by I:Scintilla)
- Syntenic dotplot: Capsella rubella - Arabidopsis lyrata
- Linum usitatissimum (common flax; linseed): http://genomevolution.org/CoGe/OrganismView.pl?oid=36226
- Syntenic dotplot: Flax -Poplar
- Gossypium raimondii (cotton): http://genomevolution.org/CoGe/OrganismView.pl?oid=36239
have both been added to iPlant CoGe. Head over and check them out. <-- But remember these genomes are protected by Fort Lauderdale for the next twelve months or until you see the genome paper.
Are we missing plant genomes you'd like to be studying? Let us know!.
iPlant User Management System Update
Dec. 18th 2011
The Data security model of CoGe has been updated. This includes creating CoGe Groups which permits the creation of user groups. These user groups may access a private set of genomes that is not accessible to other users of CoGe.
To use this, you will need to create an account with iPlant in order to be a registered CoGe user:
Major CoGe Update (version 4)
Dec. 4th 2011
Work is nearing completion for a new version of CoGe. While there are many minor improvements, additions, and changes to the tools, the major improvements are on the backend of the system including:
- New server hosted by iPlant: This means that the primary CoGe server will be located at the University of Arizona, Tucson
- Vastly expanded storage to hold even more genomes
- Enables the storage of metagenomes (as those datasets can be quite large)
- Modularized installation and centralized configuration: permits the rapid deployment of custom versions of CoGe (for those that may want a version of CoGe specific to their group of organisms)
- Federation with iPlant's authentication system:
- People will iPlant login credentials can log into CoGe as a registered user.
- Will enable the creation of personal data in CoGe
- Will enable more customization and saving of preferences for various tools in CoGe
- Will enable users to save particular analyses and datasets within CoGe
- Will enable import and export of data from CoGe to people's iPlant Data Store accounts
- Enhanced data security model:
- Will enable unpublished data to be restricted to a user or a group of users
Please come test the new CoGe: http://coge.iplantcollaborative.org and send Eric Lyons any problems you come across.
Since the holidays are coming and usage of CoGe tends to decrease, hopefully any bugs won't affect too many people while they are fixed. The migration of the domain names registered to CoGe will change once the server has been reasonably tested. Other CoGe services will migrate after that (e.g. this wiki).
CoGe domain names:
Pigeon-pea genome (Cajunus cajan) has been added to CoGe
Nov. 29th, 2011
The International Initiative for Pigeonpea Genomics has released the pigeon pea genome.
The pigeon-pea genomes may be accessed in CoGe: http://genomevolution.org/CoGe/OrganismView.pl?oid=34028
Please see this link for a syntenic dotplot between pigeonpea and medicago: http://genomevolution.org/r/49ua This syntenic dotplot has the syntenic gene pairs' evolution distance colored to differentiate orthologous and out-paralogous syntenic regions.
NCBI Genome Update: Over a thousand new genomes available in CoGe
Nov. 28th, 2011
The NCBI genome loading program for CoGe has been updated as is currently adding thousands of genomes from NCBI. Keeping CoGe current with all of the genomes at NCBI has been a challenge as their underlying data model for storing and organizing genomes evolves. The new program crawls all of NCBI's BioProjects searching for those with genomes and associated sequence. Prior to this data load there were approximately 12,100 genomes from 10,600 organisms. Approximately 40% of NCBI's BioProjects have been crawled and the current genome stats are:
Organisms: 12,093
Genomes: 13,969
Nucleotides: 305,480,720,992
Genomic Features: 99,814,749
Annotations: 224,582,292
For those that are curious, CoGe has maintained a MySQL DB transaction rate of 2000-3000 per second (majority writes/inserts) for the past 24 hours, thanks in no small part to its SSD configuration.
Note: After more performance monitoring, peak DB transactions top 9000 per second during heavy use from the genome loading programs and website activity.
Optical fun with CoGe
Nov. 22nd, 2011
Which direction does the DNA spin? Depending on how your mind is interpreting the dark and light colored dots of the DNA molecule as being "near" or "far", the helix can spin in both directions.
Thanks to Don McCarty for pointing this out.
Lamprey, Anole, and Frog genomes added/updated to CoGe
Nov. 19th, 2011
[www.ensembl.org Ensembl] version 64 genomes of Lamprey, Anole, and Frog have been added to CoGe:
Petromyzon marinus (lamprey): http://genomevolution.org/CoGe/OrganismView.pl?oid=30737 Xenopus (Silurana) tropicalis (western clawed frog): http://genomevolution.org/CoGe/OrganismView.pl?oid=33964 Anolis carolinensis (green anole): http://genomevolution.org/CoGe/OrganismView.pl?oid=33828
Both the unmasked and Masked versions of the genomes are available. For an example Syntenic dotplot between Xenopus and Tetraodon (pufferfish), please see: http://genomevolution.org/r/48w9
This dotplot uses the Syntenic path assembly to order and orient the contigs of Xenopus to the well assembled genome of Tetraodon (Frog versus Pufferfish): http://genomevolution.org/r/48w9
This dotplot uses the Syntenic path assembly to order and orient the contigs of Xenopus and Anolis (Frog V Green Lizard): http://genomevolution.org/r/48zk
Thanks to Bill Spollen for requesting these genomes.
Updated and New Plant Genome Resources
Nov. 10th 2011
The CoGePedia Sequenced plant genomes page has been updated with the latest published genomes, including the just published genomes of both pot and pidgeon pea! In addition, we have added two new pages that may be of interest to those who (like me) are constantly having to pull together introduction sections and can't remember what the right citation for well known genomic information is:
- Plant Genome Papers lists the papers describing every published plant genome, when and where it was published, and how much attention (in the form of citations) the various genomes have attracted so far.
- Plant paleopolyploidy is a list of known ancient whole genome duplications among the various plant species with sequenced genomes including information on when and how the whole genome duplications were discovered.
Both pages are clearly works in progress so please continue to contact us if we've missed genomes, whole genome duplications, or citations which should be on the list.
Main CoGe Database is down
Nov. 3rd 2011
7:00 (PCT USA) 14:00 (GMT)
Last night I ran a repair table on the main database for CoGe. This apparently ran into some problems and failed. I am currently hunting down the problem, and the main CoGe site is currently off-line. Technically, the tools are all available, but some of them are not working. The problem appears to be located in the "locations" table of the [CoGe database]. This table records the locations for all of CoGe's Genomic features. For anyone that needs to get some work done with CoGe, they are welcome to use the development server hosted at:
http://coge.iplantcollaborative.org
This version of CoGe has been under development to federate CoGe's user authentication system with the authentication system provided by the iPlant Collaborative. As such, there has been many code changes dealing with registered users and accessing restricted/private genomes. These changes are NOT fully tested and may cause some problems. Also, the development server is using an out-of-date version of the main CoGe database (though most of the genomes should be there). If you use the development server and run into any of these problems, please feel free to send Eric Lyons an email. I'd appreciate the reporting of any bugs as well as your patience with the current situation.
In case of catastrophic failure of the main database, please know that in addition to the development server, there is a full backup of the main CoGe database. These are generated weekly.
Also, thanks to Ben Field for notifying me of the problem. I deeply appreciate the help of community members in alerting me to problems with the site as well as suggestions for making it better.
Update: 8:00am
- Another "repair table" is being run on the main CoGe Database.
- Backup database is being restored on the dev server for CoGe (coge.iplantcollaborative.org). Once this is up and running, I'll point the main CoGe site to use this database and database server in case the main database has not yet been repaired.
Update: 9:30am
- backup coge database has been deployed to CoGe development server, currently undergoing "optimization" (want to avoid whatever happened to the main database)
Update: 5pm
- main coge database has been repaired. Warning and update messages taken down from the website. Let me know if anyone has any problems.
CoGe Tutorial Published in Maydica:
Oct. 24th 2011
A comprehensive open-access tutorial on using CoGe has been published in Maydica: http://www.maydica.org/articles/56_183.pdf
Abstract:
Of all the major plant groups, the grasses, with the complete genomes of five species, are the best positioned to take advantage of comparative genomics to obtain insight into functional genetic elements. Of all the grasses, maize is the best characterized in terms of genetics, development, and evolution. We provide several examples of how the web-based comparative genomics system CoGe may be used to aid in the interpretation of the maize genome sequence. These examples include verifying gene models, identifying differences between genome as- semblies, identifying conserved non-coding sequences, identifying syntenic regions between species and poly- ploidies, and identifying homeologs within maize and orthologs between maize and other grass genomes. In addition, a comprehensive list of orthologous gene sets is provided between maize and Sorghum, foxtail millet, rice, and Brachypodium.
While the article focuses on the maize genome as its primary genome, the methods are applicable to any genome.
Correction to the Classical Maize Gene and Syntelog List
Sept. 29th 2011
Phil Stinard identified an error in incorrectly assigning classical maize genes as being present in B73. Thanks to Mary Schaeffer for passing along this information and James Schnable for correcting these in the Classical Maize Gene and Syntelog List.
The following genes are now assigned as being not present in the B73:
- S
- lc1
- sn1
- hopi1
New options in SynMap
Sept. 12th, 2011
There are a couple of new options available in SynMap:
Force dotplot to be a square: You can find this option under the "Display Options" Tab with the line "Dotplot axes relations".
SVG Version of the Dotplot: There will be a new file, "SVG Version of the Syntenic Dotplot" to download in the "Links and Downloads" section of the results. This file will only appear if some form of synonymous rates are calculated and visualized (available under the "Analysis Options" tab").
Thanks to James Schnable for creating the SVG program for SynMap!
Potato genome added to CoGe
Sept. 3rd, 2011
Genome published: http://www.nature.com/nature/journal/v475/n7355/full/nature10158.html
The genome added was doubled the monoploid S. tuberosum Group Phureja clone DM1-3 516R44 (DM):
- unmasked: http://genomevolution.org/CoGe/OrganismView.pl?dsgid=12277
- masked: http://genomevolution.org/CoGe/OrganismView.pl?dsgid=12278
Please note: this version of the genome does not have annotations available.
Thanks to Will Spooner for the notification!
Brassica rapa genome added to CoGe
Sept. 3rd, 2011
Genome published: http://www.nature.com/ng/journal/vaop/ncurrent/full/ng.919.html#/group-1
Sequenced by: BGI
Brassica rapa has had a hexaploidy event subsequent to the most recent tetraploidy event in the Arabidopsis lineage.
Thanks to Will Spooner for the notification!
Cannabis sativa Pseudoassembled genome added to CoGe
Aug. 23rd, 2011
SynMap has the option to assembled one genome against another using syntenic. Such Syntneic path assemblies may be used to create a Pseudoassembly of a genome when only a contig level assembly exists. SynMap makes generating these Pseudoassemblies easy to do. Such a Pseudoassembly of the 175,000 Cannabis sativa genome was performed against the peach genome (read here to learn why peach was chosen). This pseudoassembly was reloaded back into CoGe and permits using CoGe's tools to compare the Cannabis genome at multiple levels of resolution.
To see this example: Cannabis sativa cultivar Chemdawg (marijuana)
Pseudoassemblies may be quite useful as more genomes are sequences on the cheap. Such sequencing project yield low-quality draft genomes that are usually assembled into several tens of thousands of contigs, and pseudoassemblies permit the rapid generation of large sequences that are easier to use in comparative genomic analyses.
Cannabis sativa cultivar Chemdawg (marijuana) added to CoGe
Aug. 22nd, 2011
The genome of the extremophile Cannabis sativa cultivar Chemdawg (marijuana) has been added to CoGe: http://genomevolution.org/CoGe/OrganismView.pl?oid=33804
This genome was sequenced by Medicinal Genomics (located in the Netherlands). It was sequenced with one lane of the Illumina HiSeq (2x100) platform and assembled with CLCbio’s workbench. Additional information about the assembly and genome may be found: http://www.medicinalgenomics.com/the-c-sativa-genome/
You can access Cannabis in CoGe: http://genomevolution.org/CoGe/OrganismView.pl?oid=33804
Cannabis is a member of the plant order Rosales. Of sequenced genomes in that order, the peach genome is a fantastic comparator. The reason for this is due to its high-quality sequence and assembly, and its genomic evolutionary history that does not contain any whole genome duplication event subsequent to the Eudicot paleohexaploidy shared by nearly all dicots (at least the eurosids and the astrids). As such, its genome structure is probably very similar to the common ancestor of order Rosales, and perhaps the eudicots as a whole. This likely ancestral state of the peach genome makes it quite suitable for generating a Pseudoassembly of highly fractured, low quality genome assemblies such as this Cannabis genome. CoGe's tool SynMap has an algorithm to tile contigs along any other "reference" genome in CoGe.
The Syntenic path assembly of Cannabis to the peach genome may be viewed: http://genomevolution.org/wiki/index.php/Syntenic_path_assembly#Cannabis_sativa_.28marijuana.29_v._Prunus_persica_.28peach.29
This shows the Cannabis genome sequence contains nearly the entire gene content of Peach.
Eutrema parvulum (Thellungiella parvula) added to CoGe
Aug. 17th, 2011
The genome of the extremophile crucifer Eutrema parvulum (Thellungiella parvula) has been added to CoGe: http://genomevolution.org/CoGe/OrganismView.pl?dsgid=12242
You can read about this genome in this Nature Genetics Letter: http://www.nature.com/ng/journal/vaop/ncurrent/full/ng.889.html
For a syntenic dotplot between it and Arabidopsis thaliana, please this SynMap anlaysis: http://genomevolution.org/r/3ws0
New Version of Setaria italica (foxtail millet) added to CoGe
Aug. 16th, 2011
Version 2.1 of Setaria italica has been added to CoGe. This genome was obtained from JGI/phytozome: http://phytozome.net
Unmasked version: http://genomevolution.org/CoGe/OrganismView.pl?dsgid=12240 Masked version: http://genomevolution.org/CoGe/OrganismView.pl?dsgid=12241
Thanks to Gina Turco for the request.
New Version of Fragaria vesca (woodland strawberry) added to CoGe. This time with gene models!
Aug. 11th, 2011
Version 1.1 of Fragaria vesca (woodland strawberry) has been added to CoGe http://genomevolution.org/CoGe/OrganismView.pl?dsgid=12186 .
This version contains gene models with permits more fun with syntenic dotplots: http://genomevolution.org/r/3wdb
This dotplot is strawberry versus peach. Besides from be a great summer fruit salad, this dotplot colors syntenic gene pairs based on their synonymous mutation values. From it, it is easy to see neither genome has had an independent whole genome duplication since the Eudicot paleohexaploidy event.
Thanks to Aaron Liston for requesting this genome.
Daphnia pulex (common water flea) added
Aug. 3rd, 2011 You can get all your water flea genomics here: http://genomevolution.org/CoGe/OrganismView.pl?oid=33760
Thanks to Mike Freeling for the request.
Several bugs fixed as a result of the code update
July 29th, 2011
Additional bugs were squashed today due to the major code update to CoGe's internal services. Part of the update included further modularization of the web-services from backend services. A few programs the ancillary support programs for CoGe's web-services were not correctly being passed the base configuration file for a given web-deployment and were therefore crashing. This has been corrected, but please email Eric Lyons if any problems are encountered.
Update to GenomeList
July 29th, 2011
GenomeList has been updated to:
- include a link back to GenomeList for selected genomes. This is useful if a broad selection of genomes was made and needs to be refined.
- include a link to easily download a fasta file for a given genome
- include a link to coge_gff to generate a gff file of all Genomic features and annotation in a genome
- include a TinyURL link to regenerate the genome list. This link is found at the top of the genome list.
Example GenomeList link: http://genomevolution.org/r/3v8n
Major code update to CoGe
July 27th, 2011
CoGe has undergone a major update of its web-based system today. A few bug fixes and feature enhancements mixed in, with the major one being the addition of GenomeList for creating a list of genomes, getting an overview of their genomic content, and then sending the list to other tools (e.g. CoGeBlast).
Behind the scenes was a further modularization of the web-interface from the backend support services and modules. The primary reason for this is to enable to creation of multiple CoGe installations. There has been a few requests by people for a clade/group of organisms specific installation of CoGe. With iPlant's cyberinfrastructure support, this should be possible (providing the code-base supports it).
There were some sticking points this morning migrating server specific changes from the iPlant development server to the main CoGe server, but hopefully this didn't affect too many people. However, there is a high-likelihood of additional bugs in the system that I failed to catch! Please email Eric Lyons if you find any problem.
Otherwise, we are hoping to make a full migration to iPlant's resources in the near future. iPlant's coge server is being upgraded with some additional attached storage for continual growth of the platform.
Weill's Date Palm genome version 3 has been added to CoGe
July 4th, 2011
You can find its genome in OrganismView: http://genomevolution.org/CoGe/OrganismView.pl?dsgid=11942
And a Syntenic path assembly to rice here: http://genomevolution.org/r/3ox8
This is a very rough genome (50,000+ contigs; the largest is 470KB; 13 larger than 300KB). However, the syntenic path assembly in SynMap with the option to remove any contig that doesn't have a syntenic signal makes identifying sytnenic regions a breeze (see the above link).
See this example of micro-synteny as seen in GEvo: http://genomevolution.org/r/3oxa
Thanks to: Haibao Tang, Devin O'Connor, and Jim Leebens-Mack for requesting this genome.
July 14th, 2011
The masked version of the Palm genome has been created and added to CoGe: http://genomevolution.org/CoGe/OrganismView.pl?dsgid=11947
Thanks to Haibao Tang for providing the masking procedure.
JGI's Eucalyptus grandis BRASUZ1 has been added to CoGe
You can find its genome here: http://genomevolution.org/CoGe/OrganismView.pl?oid=33537 (masked and unmasked sequence)
With a comparison to the peach genome, Eucalyptus looks to have had its own whole genome duplication subsequent to the Eudicot paleohexaploidy: http://genomevolution.org/r/3ol1
Thanks to Josquin Tibbits for recommending this genome!
Arabidopsis thaliana resequenced genomes (C24, Bur-0, Kro-0, Ler-1) from 1001genomes.org has been added to CoGe
June 30th, 2011
The "High Quality" sequences generated by the 1001genomes project for the resequencing of several arabidopsis strains has been added to CoGe. This includes:
While these genomes contain many contigs, CoGe's Syntenic path assembly algorithm can arrange and orient them against the reference genome Col-0: http://genomevolution.org/r/3okf
Thanks to Maggie Woodhouse for this suggestion!
OrganismView's Feature List display updated
June 22nd, 2011
OrganismView has a minor update for where the lists of Genomic features are displayed. The old version would display the summary list of genomic features below all the information panels. This would mean that each time a summary list was generated, it would replace the prior one. For example, if you retrieved the list first for the entire genome and second for a particular chromosome. Now, each information panel's genomic feature list appears to the right of the information summary. This allows the entire genome's feature list to be display simultaneously with the chromosome's feature list.
Broad Institute's Coccidioides group Database added to CoGe
June 21st, 2011
The entire set of sequences and associated annotations for Coccidioides has been added to CoGe. These soil fungi are pathogenic and can cause coccidioidomycosis, aka valley fever, in humans. The original data may be obtained from: http://www.broadinstitute.org/annotation/genome/coccidioides_group/MultiHome.html
And accessed through OrganismView: http://genomevolution.org/CoGe/OrganismView.pl?org_desc=Coccidioides
Thanks to Marc Orbach for suggesting and locating these genomes.
UC Berkeley Data Center Back Online
June 12th, 2011
The UC Berkeley Data Center power upgrade went smoothly. CoGe has booted up and is back online.
Thanks to:
- James Schnable for being on duty to bring CoGe down and back up.
- The entire team at the UC Berkeley Data Center for completing such a complicated upgrade to their Center and for continually updating their clients as to the progress of the operation.
CoGe Downtime June 12th, 2011
June 3rd, 2011
CoGe will be down on June 12th due to maintenance on the power infrastructure at the UC Berkeley Data Center. We will do our best to bring CoGe back up as soon as possible.
Here is their announcement:
Description: The [UC Berkeley] campus data center has been a valuable resource for campus computing for the past seven years. Demand for this highly secure, highly available, and network-redundant facility continues to rise. The current facility has reached its power and cooling capacity and Capital Projects has initiated a major renovation project intended to increase each of these capacities, while also integrating newer, more efficient systems to help the campus achieve its long-term energy conservation goals.
As part of this effort, the replacement of some core components of the data center’s power infrastructure is required. For safety reasons, a full power outage to the data center is scheduled for Sunday, June 12, 2011, from 7:00 am to 3:00 pm. The data center will rely entirely on outside air, rather than air conditioning, to provide cooling for the duration of this period. A minimal number of systems with broad campus impact, including CalMail, CalAgenda, and the campus home page, will be provided with temporary power during this outage. In the unlikely event that the data center air temperature exceeds a level appropriate for the safe operation of equipment, some of these systems may need to be shut down as well.
The list of widely used systems that are intended to remain available is below. This list is still being finalized, so additional systems may be added as campus needs require. This list will not include systems for which departments have made separate arrangements.
Citrus genomes added
May 6th 2011
The genomes of:
- Citrus clementina (Clementine mandarin): http://genomevolution.org/CoGe/OrganismView.pl?oid=33274
- Citrus sinensis (citrus, Sweet orange): http://genomevolution.org/CoGe/OrganismView.pl?oid=33273
Have been added to CoGe. These were sequenced by JGI.
A quick syntenic analysis of sinensis to peach shows that it appears to have no subsequent whole genome duplication event to the eurosid Paleohexaploidy: http://genomevolution.org/r/2zdv
Sequenced Plant Genome Phylogeny Update
May 6th 2011
James Schnable has updated the phylogeny of angiosperms for sequenced plant genomes.
CoGe Workshop at Berkeley
Apr. 19th 2011
Here is the outline/syllabus of the workshop help at Berkeley hosted by the iPlant Collaborative, the Department of Plant and Microbial Biology, QB3-CGRL (Computational Genomics Resource Laboratory), ARS-Plant Gene Expression Center, and the Freeling lab: 2011 Berkeley Workshop
This outline contains links to specific analyses used in the workshop.
Horizontal Genome Transfer
Mar. 31st 2011
Here is a fun example of a mitochondria genome being inserted into a plant chromosome: Horizontal transfer of mitochondria genome: Horizontal transfer of mitochondria genome
Second "Run GEvo Analysis!" button added to GEvo
Mar. 29th 2011
For those times when scrolling to the top of the screen to find the "Run GEvo Analysis!" button is too much work, a second button has been added at the bottom of the configuration box. This is quite useful when comparing >6 genomic regions.
Thanks to David Braun for this suggestion!
Bug Fix in FeatView
Mar. 29th 2011
Thanks to Damon Lisch for pointing out a bug in FeatView that was exposed by Firefox v4. This bug was also affecting Google Chrome (but not Safari). Please let Eric Lyons know of any problems you have running Firefox v4 (or other problems in general).
New tutorial for performing genomic rearrangement analyses
Mar. 11th 2011
A new tutorial has been written for showing how to figure SynMap to generate a link to GRIMM (by Glenn Tesler, University of California, San Diego) for performing genomic rearrangement analysis.
Tutorial: How to perform a genomic rearrangement analysis
SynMap now has support for BlastP
Mar. 7th 2011
You can now select to compare protein sequences between genomes with annotated protein coding features (CDS).
Thanks to Angelique D'Hont for the suggestion.
Cochliobolus heterostrophus C5 from JGI loaded into CoGe
Mar. 2nd 2011
You can find Cochliobolus heterostrophus C5 in OrganismView: http://genomevolution.org/CoGe/OrganismView.pl?dsgid=11258
Both masked (by JGI) and unmasked version of the genome are available.
For a syntenic dotplot between C. heterostrophus to Pyrenophora tritici-repentis strain Pt-1C-BFP (the closest relative I could find in CoGe) please follow: http://genomevolution.org/r/2m0n
This is a neat syntenic dotplot showing extensive synteny and intrachromosomeal rearrangements (though these are both contig level assemblies).
Thanks to Daniel Lawrence for request.
Sort chromosomes by name in SynMap
Feb. 26th 2011
After a couple of requests, SynMap now has an option to sort chromosomes by name instead of by size. You can read how to set this option here.
Thanks to:
- Angélique D'Hont from CIRAD
- James Schnable from UC Berkeley
for this suggestion.
How to load genomes into CoGe
Feb. 22nd 2011
If you have a CoGe installation, access to the main CoGe server, or just curious to know what is needed to load a genome into CoGe, here is a page on How to load genomes into CoGe. This is all run from the command line, and when CoGe's user permission data management system matures, this procedure will be made available via the web.
Giant Panda genome loaded into CoGe
Feb. 19th 2011
You can see the genome in CoGe at: http://genomevolution.org/CoGe/OrganismView.pl?dsgid=11227
This was one of the first big genomes sequenced using only Next Generation Sequencing Technology and assembled De novo. As a result, the assembly is rather poor compared to a fully assembled genome like the dog genome. However, through comparative genomics with SynMap, identifying syntenic regions and determining that nearly full coverage was obtained is as easy as a few mouse clicks: syntenic path assembly of the WGS panda genome to the fully sequenced dog genome. This will be quite useful as more and more large genomes are sequenced using these techniques (fast, cheap, and still very useful!)
First Metagenome loaded into CoGe
Feb. 19th 2011
Technically, there is no reason why CoGe can't store metagenomes. Its core data model stores a collection of sequences that, thus far, has been organized into a genome, but can accommodate any collection of sequences. So the first metagenome was loaded into CoGe from NCBI:
Mine drainage metagenome, whole genome shotgun sequence
And can be seen in CoGe: http://genomevolution.org/CoGe/OrganismView.pl?oid=32988
Assembling contig-level assembles to a reference genome using synteny
Feb. 18th 2011
SynMap has an option for generating a Syntenic path assembly with the click of a button. When complete, there is an option to print out your assembled genome.
CoGe 2011 Plant and Animal Genome conference presentations available for download
Feb. 10th 2011
For a complete list of PAG sessions: http://www.intl-pag.org/19/19-workshops.html
"CoGe: Comparative genomics made easy!"
Eric Lyons, iPlant Collaborative and University of Arizona, Tuscon AZ (ericlyons@e-mail.arizona.edu)
PDF available at: http://genomevolution.org/CoGe/data/distrib/presentations/PAG-2011-CoGe-CompG.key.pdf
"10,000 Genomes at Your Fingertips"
Eric Lyons, iPlant Collaborative and the University of Arizona, Tuscon AZ (ericlyons@email.arizona.edu)
PDF available at: http://genomevolution.org/CoGe/data/distrib/presentations/PAG-2011-CoGe-ComputerDemp.key.pdf
Chocolate genome gene models added
Feb. 4th 2011
Thanks to CIRAD for sharing their cacao gene models. These have been added to the Theobrama cacao genome in CoGe: http://genomevolution.org/CoGe/OrganismView.pl?dsgid=10997 .
For an example of how these gene models may be used in whole genome comparisons, see this analysis between chocolate and peach: Chocolate-peach syntenic dotplots. It shows how the evolutionary distance between sytnenic gene pairs may be visualized to differentiate between Orthologous syntenic regions derived from the divergence of these lineages, and Out paralogous syntenic regions derived from their shared Paleohexaploidy ancestry.
Arabidopsis thaliana TAIR version 10 has been added!
Jan. 27th 2011
Version 10 of the Arabidopsis thaliana genome has been added to CoGe: http://genomevolution.org/CoGe/OrganismView.pl?dsgid=11022
Thanks to all the work by the folks at TAIR
For a syntenic dotplot of version 9 versus version 10 of Arabidopsis thaliana (with the evolutionary distances of syntenic gene pairs calculated) see: http://genomevolution.org/r/2hiz
Chocolate genome added: from the International Cacao Genome Sequencing Consortium
Jan. 26th 2011
The genome of Theobroma cacao has been published: http://www.nature.com/ng/journal/vaop/ncurrent/full/ng.736.html
You can view this genome in CoGe at: http://genomevolution.org/CoGe/OrganismView.pl?dsgid=10997
To view some Syntenic dotplots of Cacao: Cacao syntenic dotplots
Of note, this genome has not had any whole genome duplication events since the Paleohexaploidy event at the base of the eurosids.
Version 2 of the Maize Genome, Now With Gene Models
Both the 50x super masked and unmasked versions of the B73_refgen2 maize genome are now updated with the new gene models released by maizesequence.org over thanksgiving break. The new genome annotation consists of 110,028 genes, many with alternative transcripts, which can be broken down as followes:
- 29,082 transposon related genes
- 17,615 putative pseudogenes
- 63,276 "real" genes. Please note while these genes were annotated as "protein coding" in the current release, they include predicted microRNA genes.
Maintenance Complete
Sept. 16th 2010
CoGe's servers have successfully be moved to a new rack space. Thanks to James, Bao, and Brent for making this happen.
Pending CoGe Maintenance
Sept. 15th 2010
We have received word from the UC Data Center which houses CoGe that we need to move our servers to a new rack space. This should only take an hour or two. Our tentative schedule time for the move is:
Sept 16th 2010 at 1pm (PCT)
We apologize for any inconvenience this may cause any of CoGe's users.
SynMap update
Aug. 26th 2010
Organisms selected in SynMap have links in their taxonomic descriptions. If you click on a term in the taxonomic description, that term is automatically entered into the organism description search. All organisms with a matching taxonomic term will be displayed. This makes it faster to find organisms related to the one in which you are interested.
OrganismView update
Aug. 26th 2010
OrganismView now has more links for finding information about an organism, and to internal CoGe tools.
External searches under organism information:
- NCBI
- Wikipedia
Internal CoGe links:
- CodeOn: automatically generates a table of amino acid usage as a function of the GC content of CDS sequences.
- SynMap: (under Genome information) automatically loads SynMap with both genomes specified to the one selected. This makes is quick to start generating whole genome comparisons and Syntenic dotplots.
Home Page update
Aug. 26th 2010
CoGe's homepage menu "Latest Genomes" now has links to search for the organism name in
- CoGe's OrganismView
- NCBI
- Wikipedia
This makes it quicker to find information on an organism, specifically if you have no idea what it is. Helpful considering that there are nearly 9,000 organisms in CoGe.
CoGeBlast update
Aug. 26th 2010
CoGeBlast now has support for specifying blastn, tblastx, lastz, megablast, and discontinuous megablast when searching with nucleotide sequences.
10,000th genome loaded!
Aug. 4th 2010
Brassica rapa has been added to CoGe and represents the 10,000th genome loaded in CoGe. Its sequence was generated by the BGI located in China. This relative of Arabidopsis is a wonderful addition to sequenced plant genomes. Their lineage share a series of whole genome duplication events (commonly known as alpha, beta, and gamma -- the latter happening prior to the radiation of the eudicots). Since their divergence, Brassica rapa has had a triploidy while Arabidopsis has had none.
Genome update from NCBI
June 28th 2010
A new update of genomes from NCBI has finished. This includes genomes from all domains of life. CoGe now has genomic sequence from 8,872 organisms comprising 9,999 genomes. There is also a new option on the homepage to list the most recently added genomes.
SIP 2010 workshop syllabus
June 23rd 2010
The syllabus for a day-long workshop on how to use CoGe for the Society for Invertebrate Pathology's conference (SIP 2010) is now available. This workshop focuses on:
- Getting an overview of how CoGe is designed for allowing scientists to create their own open-ended analyses
- Learning what the various tools in CoGe do and they to use them
- Working through specific sets of example problems focused on analyzing two groups of organisms important for invertebrate pathology: baculoviruses and Bacillus thuringiensis
The workshop's syllabus is available: SIP2010
CoGe's update progress
June 18th 2010
The switch to the new server went as smoothly as I could have hoped.
Besides from new hardware (which should greatly accelerate many of CoGe's analyses and improve system stability), this installation welcomes a new version of CoGe too!
This new version of CoGe has:
- Update UI
- Various feature extensions on existing tools
- Updated algorithms (new blast API with support for the megablast families, LastZ)
- New database additions
- Update of core modules for database API
- New configuration files that will help deployment of CoGe to new sites
Please contact Eric Lyons if you find any bugs!
Today is the day
June 17th 2010
Going to through the switch today. Expect some downtime with CoGe and some support systems being temporarily off line.
New CoGe Server Update
June 10th 2010
It appears that most of the software updates and migration to the new server are working. We have deployed the new server to the UC Data Center, but due to some complications with rack-space, IP address allocation, sub-nets, firewalls, etc., things may be in flux for a while. We've had to take our development server (aka toxic) off line and put the new server on its IP address till those things get sorted out. In the meanwhile, we will plan on making the switch to production on the new server soon (hopefully next week). When this happens, expect CoGe to be offline for a couple of hours, but we will do our best to keep downtime to a minimum.
New CoGe Server is being readied!
June 2nd 2010
We have our new server for CoGe! Its deployment will not only include new performance improvements due to more computing power, but all several changes and additions to CoGe:
- new user interface
- new algorithm options
- new structure of the underlying code-base to make it is easier to redeploy (in anticipation of eventually getting the code-base released to those interested)
We are planning on moving the new server to the UC data center this Fri. After some more testing and bug hunting, we will switch our current production server's IP address to this machine. There is a high chance that there will be some downtime for CoGe during this switch and we will post announcements as to when this change will happen! In the meanwhile, if anyone is interested in testing new CoGe, please e-mail Eric Lyons.
SGRP: (Sanger Institute) yeast genomes added to CoGe
May 18th 2010
75 Yeast genomes from SGRP (Saccharomyces Genome Resequencing Project) have been added to CoGe. For a complete list of Organisms, please see SGRP: Sanger Institute Yeast Genomes.
CoGe post on The OpenHelix
May 5th 2010
Eric Lyons wrote a piece about CoGe for The OpenHelix Blog
Version 2 of Maize B73 genome added to CoGe
May 3rd 2010
This release does not yet have annotations (yet)!
You can view the genome in CoGe at: http://genomevolution.org/CoGe/OrganismView.pl?dsgid=9106
This sequences was obtained from: http://www2.genome.arizona.edu/genomes/maize
And can read about differences in the assembly between versions 1 and 2: here.
Version 2 of Vitis vinifera (grapevine) genome added to CoGe
Apr. 10th 2010
You can view the genome in CoGe at: http://genomevolution.org/CoGe/OrganismView.pl?dsgid=9048
Version 2 with 12x coverage was obtained from Genoscope.
There are some changes to the assembly with new contig orders and additional sequence added to the pseudomolecules which can been seen here.
New NCBI Genome Update. CoGe surpasses 8,900 genomes from 8,200 organisms
Apr. 9th 2010
Finished an update from NCBI. However, this is not a complete listing of all genomes available at NCBI due to some API problems getting some genomes. You can read about this problem below.
Version 3 of Medicago truncatula added to CoGe
Apr. 9th 2010
You can view the genome in CoGe at: http://genomevolution.org/CoGe/OrganismView.pl?dsgid=8976
Syntenic comparison of version 3 to version 2 shows extensive changes in the primary sequence. Some chromosomes have had their sequence substantially updated.
Prunus persica (peach tree) added to CoGe
Apr. 9th 2010
You can view its genome in CoGe at: http://www.genomevolution.org/CoGe/OrganismView.pl?oid=30980
Its genome was produced by the International Peach Genome Initiative and its sequence was obtained from phytozome. This genome is currently unpublished and therefore under the publication restrictions of the Fort Lauderdale Convention.
Peach is a eudicot in the Rosaceae family.
Automatic NCBI Genome Loader Update
Apr. 8th 2010
The automatic NCBI genome loader is running today. It has been a while since I last ran it after running into an API problem with NCBI's eutils tools three months ago. The issue is still unresolved and even after checking in every two weeks for a status update, I have yet to receive any word as to when the bug will be fixed. For those interested, here is my bug report sent at the end of January:
Issue (http://jira.be-md.ncbi.nlm.nih.gov/browse/HD-1843): Key: HD-1843 Summary: Unable to get some genomes using eutils Type: Task Status: In Progress Priority: Normal Assignee: Matten, Wayne Reporter: Nobody Description: Hi, I've be checking which genomes are available from NCBI using eutils by getting a list of all the genome project ids (genomeprj) and then retrieving their associated genome ids. I've found that a lot of the recently deposited genomes (usually with accessions CPXXXXXX) are have a genomeprj id but no associated genome id. For example, genomeprj=30031. It is listed in this list: http://eutils.ncbi.nlm.nih.gov/entrez/eutils/esearch.fcgi?db=genomeprj&term=all%5Bfilter%5D&retmax=999999 But has no genome id: http://eutils.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?db=genome&dbfrom=genomeprj&id=30031 However, it does have an entry in genbank: http://www.ncbi.nlm.nih.gov/nuccore/CP001637.1?ordinalpos=3&itool=EntrezSystem2.PEntrez.Sequence.Sequence_ResultsPanel.Sequence_RVDocSum I am probably missing something obvious. Can you help me figure out how to get a list of all the genomes at NCBI? I am using these data in an NSF funded and publicly available comparative genomics platform (http://synteny.cnr.berkeley.edu/), and have programs that check for new genomes and new versions of existing genomes from NCBI on a periodic basis. It is important for this system to be as up to date as possible with regards to the large number of genomes that are becoming available as there are many researchers using this tool for their work. Thanks in advance for your help, -Eric Lyons
If anyone has any solutions to this problem, please contact me.
Major bug fix in SynMap
Mar. 26th 2010
While testing the prior bug fix, I discovered that SynMap wasn't working on genomic sequence comparisons (as opposed to CDS sequence comparisons). This was due to the new analytical pipeline's data processing requiring unique names for each blast hit. Otherwise, multiple hits to the same sequence name would get removed as a Local duplicate. As all hits to a genomic sequence were named according to the chromosome, all such hits were flagged as Local duplicates and removed from the analysis.
As always, if you find a problem in CoGe, feel free to email Eric Lyons and let him know what you've found. There are now too many options and buttons to click in CoGe for me to test with each update.
Minor bug fix in SynMap
Mar. 25th 2010
With SynMap's new analytical pipeline, there are still some bugs to be worked through. Hopefully got one today in the script that converted blast input files to bed format, which is required for the program to find local duplicates in the compared genomes. These local duplicates are removed from the algorithm for finding collinear series of putative homologous genes used to infer syntenic regions. Also, these local duplicate files are displayed in the download section of the results in case they are wanted for other analyses.
Hosting local tiny URL encoding
Mar. 24th 2010
Replaced using tinyurl.com for a local installation of a URL hashing and redirecting service. Makes generating these faster and allows for customized names. Note: the tinyurls will still work.
Sequenced plant genomes
Mar. 13th 2010
James Schnable has created a page detailing all of the sequenced plant genomes including:
- overview of their genomic content
- publications
- status of completion
- interesting factoids (e.g. The average US American eats 25lbs of bananas a year.)
Read about them here: Sequenced plant genomes
The JGI's Manihot esculenta (cassava) genome has been added
Mar. 13th 2010
This genome from the JGI brings CoGe up-to-date with phytozome v5.0.
You can access cassava in CoGe here, and get more information from phytozome.
The JGI's Cucumis sativus (cucumber) genome has been added
Mar. 12th 2010
You can access it in CoGe here. Or get more information about it from phytozome. This is apparently a distinct sequence from the one in Nature Genetics last November. That sequence was from "'Chinese long' inbred line 9930" this version comes from the inbred Gy14. More details here
SynMap updated
Mar. 12th 2010
After a month of work, SynMap has undergone several significant changes, incorporating new algorithms written by Haibao Tang and Brent Pedersen:
- new merging function for overlapping and neighboring diagonals (program: quota alignment)
- new method for detected tandem gene duplicates
- better reporting of all intermediate files used in the analysis, including tandem duplicates
These changes have also hoped to increase the stability of SynMap, which due to its long pipeline, has been known to crash for some genomes and/or specific parameter configurations. Please let Eric Lyons know if you have any problems with an analysis. Please send along the names of the organisms/genomes compared and a copy of the log file produced by each SynMap run (if possible).
Persistent GEvo bug fixed
Mar. 11th 2010
A long-stranding, but intermittent and annoying bug in GEvo has finally fixed. This (hopefully) solves the problem where once in a while, GEvo will return blank results to its interactive viewer, Gobe. The crux of the bug, and why it was intermittent (and hence difficult to reproduce and trouble-shoot), was a race condition between asynchronous client javascript code and server perl code. Perl was responsible for generating a random session id for the analysis, but it occasionally failed to return that id to the client code before the analysis was sent back to the server for processing. When this happened, the processing analysis received a default id and multiple analyses could be merged if the default id had been used within that past 24 hours (the length of time an analysis stays on the server before being deleted). When Gobe tried to process the results, the stored data and what was specified for initialization did not match, thus causing gobe to fail and return blank results. The solution: have javascript generate the analysis session id so there is no chance of a delay before the analysis is sent to the server for processing.
However, if anyone does come across this bug again (or any others), please let me know: Eric Lyons
Rice Version 6.1 loaded
Mar. 10th 2010
You can view it in GenomeView. This was retrieved from MSU's Rice Genome Annotation Project.
The classic set of Maize Genes
Mar. 9th 2010
James Schnable manually evaluate ~460 classic maize genes available from MaizeGDB and NCBI, determined their genomic positions in the maize genome, and found their Syntenic regions within maize (from its most recent Whole genome duplication event), sorghum, rice, and brachypodium. This list contains links to compare these syntenic regions using GEvo.
New plant genomes in CoGe
Feb. 10th 2010
Mimulus guttatus (monkey flower): http://synteny.cnr.berkeley.edu/CoGe/OrganismView.pl?oid=30760 Mimulus is an outgroup to the rosids (in the sister group, the asterids)
Populus trichocarpa (Poplar; cotton wood): http://synteny.cnr.berkeley.edu/CoGe/OrganismView.pl?oid=324 Version 2 of poplar!
Both are from the JGI.
MaizeGDB links to GenomeView
Feb. 8th 2010 MaizeGDB is now linking to CoGe's GenomeView so maize researchers can find maize-sorghum Syntenic gene sets and quickly perform syntenic analyses using GEvo. For an example view from MaizeGDB's genome broswer:
http://gbrowse.maizegdb.org/cgi-bin/gbrowse/maize/?name=chr1:1000000..1200000
For instructions on how to perform this workflow: MaizeGDB and CoGe
For more information on maize-sorghum syntenic analyses: Maize-Sorghum genome analyses
For a quick video walk through of the new connections: MaizeGDB_and_CoGe.27s_Maize-Sorghum_Orthologies
Syntelog visualization in GenomeView
Feb. 5th 2010 GenomeView has been updated to auto-detect Genomic features with annotations that are links to GEvo. These links provide an analysis of a Genomic feature (e.g. gene) to previously identified Syntologous sets of features. Currently, this has been implemented using syntelogs from maize and sorghum, but with the code in place, we will expand annotations for genomic features from other organisms for which we generated syntologous gene sets. For an example of this visualization in GenomeView please see: | this GenomeView of sorghum. Also, for an expanded list of glyphs used in GenomeView please refer to these examples.
Easy exporting and downloading of genomes
Jan. 16th 2010 OrganismView has new options for easily downloading the sequences of a genome in fasta format and retrieving all of its annotations in an GFF file. To access, just search for an organism and genome of interest, and look for the links under "Genome Information".
FastaView is linked to phylogeny.fr for one-click phylogenetics
Jan. 10th. 2010
We've linked to phylogeny.fr for quick and easy phylogenentic tree reconstruction. Now, you can build a list of fasta sequences and display them in FastaView, select protein or DNA sequences, edit them if necessary (e.g. add or remove sequences manually), and press a button to send them off to phylogeny.fr for:
- multiple sequence alignment (MUSCLE)
- maximum likelihood phylogenetic tree reconstruction (PhyML)
- tree visualization (TreeDyn)
For an example, use this link to FastaView and press the button "phylogeny.fr" at the bottom of the screen.
Special thanks to Haibao Tang for pointing out this incredible web resource!
Haibao Tang joins the Freeling lab
Jan. 4th 2010
Haibao Tang, an expert in plant comparative genomics and genome evolution, as well as a great python programmer, has joined the Freeling lab. His input and contributions will be most valued!
New Tutorials added
Jan. 4th 2010
New Tutorials have been added:
- How to find syntenic regions between genomes
- How to find inversions
- How to find rarely and frequently used codons in a genome
- How to generate an amino acid usage table for a genome
- Using synonymous mutation rates in SynMap to rapidly identify different whole genome evolutionary events
- How to extract all gene sequences from a genomic region
- How to identify putative horizontal gene transfer events
Linked to ProSite for protein domain searching
Dec. 24th 2009
FastaView is now linked to ProSite when viewing a protein sequence for protein domain searching. See this FastaView example and click on the link at the bottom of the page.
Improved implementation of DAGChainer in SynMap
Dec. 15th 2009
Thanks again to Brent Pedersen for some fantastic programming. He discovered that DAGChainer's C++ code's makefile did not include the -O3 optimization, rewrote the input/output methods of the compiled binary to read from STDIN instead of a file, and rewrote the perl front-end in python. Together, these changes increase CoGe's DAGChainer implementation in SynMap between 2-4 fold.
You can download his code at: svn co http://bpbio.googlecode.com/svn/trunk/scripts/dagchainer
CoGe Workshop being taught at SIP 2010
Nov. 30th 2009
Genomics: What every invertebrate pathologist needs to know. http://www.sip2010.org/index.php/Bioinformatics-Workshop.html
CoGe on OpenHelix and James and the Giant Corn
Nov. 18th 2009
Phillipe Lamesch from TAIR passed along a link to openhelix.com highlighting CoGe's tool GEvo. They put together a nice video showing GEvo. They, in turn, found this on a posting at the blog of James and the Giant Corn who had used GEvo for a grant proposal.
Maize Pseudomolecule Assembly with Gene Models Released
Oct. 20th 2009
Thanks to maizesequence.org for providing the sequence and annotations. The current pseudomolecule assembly of maize has been loaded into CoGe.
- Link to OrganismView for complete set of gene models.
- Link to OrganismView for filtered gene model set.
- Maize-Sorghum syntenic dotplot with syntologs colored by synonymous rate change.
- Maize-Maize syntenic dotplot with syntologs colored by synoonymous rate change.
CoGe surpasses 7000 organisms in its database!
More fun for everyone!
NCBI Genome Loader Updated
CoGe's automated NCBI genome loader has been updated and is once again checking NCBI regularly for new and updated genomes. You can get a snapshot of the number or organisms and genomic sequence in CoGe by checking its homepage, search for your genome of interest using OrganismView.
CoGe is linked to TARGeT: Tree Analysis of Related Genes and Transposons
You can send a set of fasta sequence generated by FastaView directly to TARGeT.
New version of Gobe release!
Read general announcement Gobe. Major feature: transparent wedges are drawn to connect regions of sequence similarity.
Version 3 of CoGe is released!
Read general announcement CoGe version 3.