Using CoGe for the analysis of Plasmodium spp
About this guide
This 'cookbook' style document is meant to provide an introduction to many of our tools and services and is structured around a case study of investigating genome evolution of the malaria-causing Plasmodium spp. The small size and unique features of this pathogen's genome make it ideal for beginning to understand how our tools can be used to conduct comparative genomic analyses and uncover meaningful discoveries.
Through a number of example analyses, this guide will teach users about the following tools:
- LoadGenome: Add a new genome to CoGe.
- LoadAnnotation: Add structural and/or functional annotations to a genome.
- GenomeInfo: Get information about a genome.
- GenomeList: Get information about several genomes in a table.
- CoGeBLAST: BLAST against any set of genomes.
- GEvo: Microsynteny analysis.
- SynMap: Whole genome syntenic analysis.
- - SynMap#Calculating and displaying synonymous/non-synonymous (Ks, Kn), data Kn/Ks Analysis: Characterize the evolution of populations of genes.
- - SPA tool: Syntenic Path Assembly to assist in genome analysis.
- SynFind: Identify syntenic genes across multiple genomes.
- CodeOn: Characterize patterns of codon and amino acid evolution in coding sequence.
A brief introduction to Plasmodium genome evolution
The study of parasitic genomes via comparative genomics offers many unique challenges. Parasite genomes are characterized by a combination of gene loss and the acquisition of species- or lineage-specific genes; in particular, many specialized genes mediate host–parasite interaction [1]. The dynamic nature of parasitic genomes is particularly evident within the genus Plasmodium. The genus emerged ~40 million years ago and harbors roughly 200 species of parasitic protozoa better known as malaria parasites. All Plasmodium species have a complex life cycle involving some kind of vertebrate host and a mosquito vector of the genus Anopheles (mammals) or Culex (birds). In addition, Plasmodium species share similar life cycle characteristics, albeit with a few exceptions (e.g. hypnozoites). However, host and vector preferences differ among Plasmodium species [2].
Plasmodium genomes are tiny (between 17-28Mb) in comparison to those of their vertebrate (1Gb for birds; 2-3Gb for mammals) and mosquito (230–284Mbp) hosts [3]. All Plasmodium genomes consist of fourteen chromosomes (nuclear genome), as well as a mitochondrial and apicoplast genome. Despite these shared genomic characteristics, the structural organization, gene content, and sequence of Plasmodium genomes is highly variably within the genus [4]. The exact origins and mechanisms of these differences remain largely unexplored, however, they are generally hypothesized to stem from host shift events [5][6].
An increase in funding devoted to malaria research has coincided with a dramatic increase in publicly available genomic information for Plasmodium [7]. The most prominent repository is found at NCBI/Genbank [8]; while additional and unique sequences can also be found on other databases: PlasmoDB [9], GeneDB [10], and MalAvi [11]. This wealth of genomic data facilitates detailed comparative genomic approaches, opening the possibility to:
- Infer origins of certain traits, specialized phenotypes, and genomic features.
- Track the maintenance of conserved genes across the genus, as well as the gain or loss of genes unique to a single species or a group of closely related species.
- Identify the potential historical interactions that might have lead to the development of genomic adaptations.
Through a case study on Plasmodium evolution, we will illustrate how CoGe can be used for the analysis of multigene families, local synteny, and whole genome comparisons (genome composition, rearrangement events, and gene order conservation).
Finding and integrating Plasmodium genomes in CoGe
You can find the details of Plasmodium genome integration in the following link: Finding and intregating Plasmodium genomes to CoGe
Using CoGe tools to perform comparative analyses
Workflows
The following links direct to specific tools for the comparative analysis of Plasmodium genomes:
Plasmodium analysis workflow 1: Tools that evaluate genomic properties and amino acid usage
Identifying gene homologs (CoGeBLAST)
The identification of homology based on sequence similarity is a key tool for gaining insight into an organism’s biology and genetics. Defining evolutionary relationships and inferring common ancestry is particularly challenging when dealing with multigene families. Plasmodium multigene families perform a wide array of functions, have diverse gene organization, and distinct evolutionary histories. Here we focus on a set of multi-gene families arising from the subtelomere (e.g. var, stevor, rifin, or vir) that have very complex evolutionary patterns and organizations [12]. These four gene families are of particular interest because of their role in immune evasion and cell invasion. In addition, these families have undergone rapid sequence evolution and gene turnover [13][14][15]. These factors make inferring orthology/paralogy and gene gain/loss events in Plasmodium subtelomeric families a complex task.
The 313 members of P. vivax’s vir family are grouped into 10 subfamilies based on their sequence similarity. Gene size and structure (number of exons) is largely variable among family members [16][17][18]. The genetic diversity in the vir family is larger than that of other P. vivax families. Only fifteen of the 313 vir genes are shared across all sequenced P. vivax strains despite the recent emergence of the species ~ five million years ago. Within this group, PVX_113230 has been proposed as a potential family founder based on its high sequence conservation [19].
Here we use CoGeBLAST to identify the proposed founder of the Plasmodium vir family (PVX_113230) in six P. vivax strains (including the recently sequenced PO1 strain). CoGeBLAST incorporates genome visualization into BLAST analyses. Therefore, this tool facilitates the study of complex evolutionary patterns.

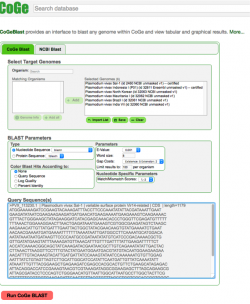
| The following steps show how to use CoGeBLAST in the CoGe platform:
2. Click on CoGeBLAST or follow this link: https://genomevolution.org/coge/CoGeBlast.pl 3. Type the scientific name of the Organism of interest in the Search box. All genomes with names matching the search term will appear under the Matching Organisms menu. Notebooks matching the term will appear in a new window after clicking on Import List. 4. Select all the genomes of interest and click on + Add. The genomes will now appear on the Selected Genomes menu. You can also select any of your Notebooks and include all the genomes contained in it. 5. Enter your query sequence in FASTA format. If desired, you can change the BLAST Parameters before starting the analysis. 6. Once all information is included click on Run CoGe BLAST (Figure 11). 7. The analysis output will include:
You can find links to the FASTA sequences used in this analysis in the "Sample data" section at the end of this page. |
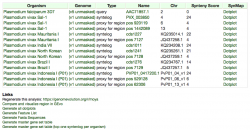
Sequences with significant similarity to PVX_113230 were found in all the evaluated P. vivax strains, including PO1. However, the number of high-scoring segment pairs for each P. vivax genome was variable. The highest number of sequence homologs was observed in the strains: Mauritania, PO1, and Salvador-1. Sequence divergence of vir members within P. vivax seems to affect the number of high-scoring segment pairs per strain. Thus, the variation in the number of HSPs across strains further supports observations about the high sequence variation among vir homologs.
The location of HSPs appears to be slightly variable across genomes. However, we cannot confirm this patterns until the Mauritania, North Korea, Brazil I, and India VII genomes are fully assembled. Between the two fully assembled P. vivax genomes (Salvador-1 and PO1), BLAST hits were located largely in the same chromosome regions (Figure 12). As expected, a higher number of BLAST hits and a more variable genome location were observed when a less conserved vir family member (PVX_096004.1) was used as a query (analysis can be run following this link: https://genomevolution.org/r/mkcg).
Identifying sets of syntenic genes amongst several genomes (SynFind)
Small-scale genomic rearrangements are often linked to species-specific gene gain/loss events. Family-linked rearrangements are observed amongst closely related Plasmodium species, and in occasion, at an intra-specific level. CoGe’s tool, SynFind, is used to identify gene homologs across any number of genomes, and thus can be of use to identify these rearrangements.
The evolutionary trajectory of multigene families can be difficult to infer, especially in those with a scattered organization or rapid gene turnover. While this issue is particularly prevalent in species-specific gene families; genus-specific families can present intricate evolutionary patterns as well. One good example can be found in the SERA (serine repeat antigen) family, a gene family that has experienced a significant number of inter-specific contractions, expansions, and rearrangements. These patterns remain to be evaluated at an intra-specific level. We will use SynFind to study family's organization of SERA paralogs in 6 P. vivax strains.
SERA paralogs are expressed during various stages of the Plasmodium life cycle. All SERA family members encode proteins with a papain-like cysteine protease motif [20]. These motifs are commonly found both inside and outside the genus Plasmodium [21][22]. One member (SERA-5), expressed during late trophozoite and schizont stages, has been considered as a promising malaria vaccine target [23]. We will use this gene sequence as a query for the SynFind analysis.
| The following steps show how to use SynFind:
2. Click on SynFind or follow this link: https://genomevolution.org/CoGe/SynFind.pl. 3. Type a scientific name of your search bar under Select Target Genomes. Organisms and genomes with names matching the search term will be displayed on the Matching Organisms menu. 4. Select the genomes of interest using Ctl+click or Command+click, then click on + Add. The genomes will appear on the Selected Genomes menu. You can also import genomes from your Notebooks. 5. Type the Name, Annotation, or Organisms on the Specify Features section. It is recommended to include as many specific terms as possible. Once done click on Search. 6. All matches to the search term and the genome where they have been found will appear in a new menu within the same section. Select all relevant Matches and the reference Genome. 7. Click on Run SynFind to start the analysis. 8. SynFind will output all syntenic regions from the reference genome and their Syntenic depth. This output can be used as a query for other CoGe tools.
GEvo results can be replicated here: https://genomevolution.org/r/mpdf |
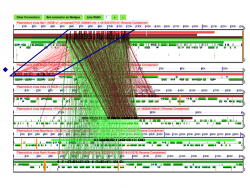
We used Synfind to identify genes homologous to SERA-5 across 6 P. vivax genomes (Figure 21). Synfind’s output was used as a query for a GEvo analysis of the region. Our results show a conserved number of SERA paralogs in all P. vivax strains. The organization of the SERA family was different on the Brazil I strain respect to other P. vivax strains (Figure 22). Previous studies on SERA have suggested that some family members are unique to P. vivax and closely related species [24]. Our results indicate that family organization is not completely conserved on the intra-specific level. This is most evident on recently duplicated paralogs.
SynFind also identified matching segments outside the SERA multigene family. These segments belonged to hypothetical protein coding genes, ATP proteases, and uncharacterized transcripts. Papain-like cysteine protease motifs are commonly found outside both Plasmodium and the SERA family. Thus, is likely that these segments share a papain-like cysteine protease motif but are not evolutionarily related to SERA.
Additional tools for genome analysis with CoGe
You can learn about the SPA usage on Plasmodium genomes in the following link: Plasmodium genome analysis using Syntenic Path Assembly
Overall conclusions
The number of available Plasmodium genomes has increased considerably during recent years. This wealth of genomic information creates an unprecedented opportunity to study the unique genomic qualities of this genus using comparative genomics.
There have been tremendous achievements in malaria treatment and control strategies. Thanks to worldwide efforts, there has been a significant reduction in the number of malaria cases and malaria-related deaths between 2000 and 2015. By 2015, it was estimated that the number of malaria cases decreased from 262 million to 214 million, and the number of malaria-related deaths from 839,000 to 438,000 [25]. However, there are still numerous aspects of malaria research that need to be further addressed.
The intricacies of parasite-host relations in Plasmodium infection might be more complex than previously considered [26]. Humans have recently been infected by Plasmodium species classically considered specific to non-human primates (e.g. a single infection with P. cynomolgi [27] and various infections with P. knowlesi [28]). In addition, african primates have been infected by unique P. falciparum strains (a parasite classically considered exclusive to humans) and are proposed to act as reservoirs for this parasite [29][30]. In bird Plasmodium, the putative evolutionary time of parasite-host associations has a significant role in the development of pathogenicity and in host mortality [31]. Finally, multiple host-switch events between largely divergent host types are thought to have occurred in bat Haemosporidia [32]. These cases highlight the complexity of the Plasmodium infection landscape. Insights into the unique patterns of Plasmodium biology, epidemiology, ecology, and genetics can be obtained from molecular and comparative genomic studies.
The rapid growth of genomic information makes implementing tools that facilitate assessing genome evolutionary trends an imperative task. The services and tools provided by the CoGe platform are of considerable use in advancing Plasmodium comparative genomics. Here, we showed how various CoGe tools could be used to assess evolutionary patterns unique to Plasmodium. We also showed how to use this platform to further characterize sequenced Plasmodium genomes. Overall, we have demonstrated that CoGe’s tools can be used to address evolutionary questions such as:
- The evolutionary origins of Laveranian AT-rich genomes.
- The location and nature of genome rearrangements between Plasmodium.
- The evolutionary patterns of genes crucial in cell invasion.
- The evolutionary trends of multigene families.
Useful links
Plasmodium Notebooks in CoGe
- Link to Notebook for published Plasmodium genome data: https://genomevolution.org/coge/NotebookView.pl?lid=1753
- Link to Notebook for published P. falciparum strains: https://genomevolution.org/coge/NotebookView.pl?lid=1758
- Link to Notebook for published P. vivax strains: https://genomevolution.org/coge/NotebookView.pl?lid=1760
- Link to Notebook for published Plasmodium apicoplast data: https://genomevolution.org/coge/NotebookView.pl?lid=1754
- Link to Notebook for published Plasmodium mitochondrion data: https://genomevolution.org/coge/NotebookView.pl?lid=1756
Sample data
- Gene sequences used on CoGeBLAST analysis (obtained from PlasmoDB):
- PVX_113230.1 | Plasmodium vivax Sal-1 | variable surface protein Vir14-related (http://plasmodb.org/plasmo/app/record/gene/PVX_113230)
- PVX_096004.1 | Plasmodium vivax Sal-1 | VIR protein (http://plasmodb.org/plasmo/app/record/gene/PVX_096004)
- Gene sequence used on SynFind to inform GEvo analysis (obtained from PlasmoDB):
- PVX_003830.1 | Plasmodium vivax Sal-1 | serine-repeat antigen 5 (SERA) (http://plasmodb.org/plasmo/app/record/gene/PVX_003830)
- Gene sequences used on CoGeBLAST to inform GEvo analysis (obtained from PlasmoDB):
- PF3D7_0424100.1 | Plasmodium falciparum 3D7 | reticulocyte binding protein homologue 5 (http://plasmodb.org/plasmo/app/record/gene/PF3D7_0424100)
- PVX_096410.1 | Plasmodium vivax Sal-1 | cysteine repeat modular protein 2, putative (http://plasmodb.org/plasmo/app/record/gene/PVX_096410)
References
- ↑ Jackson AP. 2015. Preface. The evolution of parasite genomes and the origins of parasitism. Parasitology. 142 Suppl 1:S1-5. https://www.ncbi.nlm.nih.gov/pubmed/25656359
- ↑ Sinka ME, Bangs MJ, Manguin S, Rubio-Palis Y, Chareonviriyaphap T, Coetzee M, Mbogo CM, Hemingway J, Patil AP, Temperley WH, Gething PW, Kabaria CW, Burkot TR, Harbach RE, Hay SI. 2012. A global map of dominant malaria vectors. Parasit Vectors. 5:69. https://www.ncbi.nlm.nih.gov/pubmed/22475528
- ↑ DeBarry JD, Kissinger JC. 2011. Jumbled Genomes: Missing Apicomplexan Synteny. Mol Biol Evol. 2011 Oct; 28(10): 2855–2871. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3176833/
- ↑ Carlton JM, Perkins SL, Deitsch KW. 2013. Malaria Parasites. Caister Academic Press
- ↑ Prugnolle F, Durand P, Ollomo B, Duval L, Ariey F, Arnathau C, Gonzalez JP, Leroy E, Renaud F. 2011. A Fresh Look at the Origin of Plasmodium falciparum, the Most Malignant Malaria Agent. PLoS Pathog. 7: e1001283. http://journals.plos.org/plospathogens/article?id=10.1371/journal.ppat.1001283
- ↑ Prugnolle F, Rougeron V, Becquart P, Berry A, Makanga B, Rahola N, Arnathau C, Ngoubangoye B, Menard S, Willaume E, Ayala FJ, Fontenille D, Ollomo B, Durand P, Paupy C, Renaud F. 2013. Diversity, host switching and evolution of Plasmodium vivax infecting African great apes. Proc Natl Acad Sci U S A. 110:8123-8. https://www.ncbi.nlm.nih.gov/pubmed/23637341
- ↑ Buscaglia CA, Kissinger JC, Agüero F. 2015. Neglected Tropical Diseases in the Post-Genomic Era. Trends Genet. 31:539-55. https://www.ncbi.nlm.nih.gov/pubmed/26450337
- ↑ Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. 2016. GenBank. Nucleic Acids Res. 44: D67–D72. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4702903/
- ↑ Aurrecoechea C, Brestelli J, Brunk BP, Dommer J, Fischer S, Gajria B, Gao X, Gingle A, Grant G, Harb OS, Heiges M, Innamorato F, Iodice J, Kissinger JC, Kraemer E, Li W, Miller JA, Nayak V, Pennington C, Pinney DF, Roos DS, Ross C, Stoeckert CJ Jr, Treatman C, Wang H. 2009. PlasmoDB: a functional genomic database for malaria parasites. Nucleic Acids Res. 37:D539-43. https://www.ncbi.nlm.nih.gov/pubmed/18957442
- ↑ Logan-Klumpler FJ, De Silva N, Boehme U, Rogers MB, Velarde G, McQuillan JA, Carver T, Aslett M, Olsen C, Subramanian S, Phan I, Farris C, Mitra S, Ramasamy G, Wang H, Tivey A, Jackson A, Houston R, Parkhill J, Holden M, Harb OS, Brunk BP, Myler PJ, Roos D, Carrington M, Smith DF, Hertz-Fowler C, Berriman M. 2012. GeneDB--an annotation database for pathogens. Nucleic Acids Res. 40:D98-108. https://www.ncbi.nlm.nih.gov/pubmed/22116062
- ↑ Bensch S, Hellgren O, Pérez-Tris J. 2009. MalAvi: a public database of malaria parasites and related haemosporidian in avian hosts based on mitochondrial cytochrome b lineages. Mol Ecol Resour. 9:1353-8. https://www.ncbi.nlm.nih.gov/pubmed/21564906
- ↑ Singh V, Gupta P, Pande V. 2014. Revisiting the multigene families: Plasmodium var and vir genes. J Vector Borne Dis. 51:75-81. https://www.ncbi.nlm.nih.gov/pubmed/24947212
- ↑ Niang M, Yan Yam X, Preiser PR. 2009. The Plasmodium falciparum STEVOR multigene family mediates antigenic variation of the infected erythrocyte. PLoS Pathog. 5:e1000307. https://www.ncbi.nlm.nih.gov/pubmed/19229319
- ↑ Witmer K, Schmid CD, Brancucci NM, Luah YH, Preiser PR, Bozdech Z, Voss TS. 2012. Analysis of subtelomeric virulence gene families in Plasmodium falciparum by comparative transcriptional profiling. Mol Microbiol. 84:243-59. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3491689/
- ↑ Petter M, Bonow I, Klinkert MQ. 2008. Diverse expression patterns of subgroups of the rif multigene family during Plasmodium falciparum gametocytogenesis. PLoS One. 3:e3779. http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0003779
- ↑ Carlton JM, Adams JH, Silva JC, Bidwell SL, Lorenzi H, Caler E, Crabtree J, Angiuoli SV, Merino EF, Amedeo P, Cheng Q, Coulson RM, Crabb BS, Del Portillo HA, Essien K, Feldblyum TV, Fernandez-Becerra C, Gilson PR, Gueye AH, Guo X, Kang'a S, Kooij TW, Korsinczky M, Meyer EV, Nene V, Paulsen I, White O, Ralph SA, Ren Q, Sargeant TJ, Salzberg SL, Stoeckert CJ, Sullivan SA, Yamamoto MM, Hoffman SL, Wortman JR, Gardner MJ, Galinski MR, Barnwell JW, Fraser-Liggett CM. 2008. Comparative genomics of the neglected human malaria parasite Plasmodium vivax. Nature. 455:757-63. https://www.ncbi.nlm.nih.gov/pubmed/18843361
- ↑ Lopez FJ, Bernabeu M, Fernandez-Becerra C, del Portillo HA. 2013. A new computational approach redefines the subtelomeric vir superfamily of Plasmodium vivax. BMC Genomics. 14:8. https://www.ncbi.nlm.nih.gov/pubmed/?term=A+new+computational+approach+redefines+the+subtelomeric+vir+superfamily+of+Plasmodium+vivax
- ↑ Fernandez-Becerra C, Yamamoto MM, Vêncio RZ, Lacerda M, Rosanas-Urgell A, del Portillo HA. 2009. Plasmodium vivax and the importance of the subtelomeric multigene vir superfamily. Trends Parasitol. 2009 25:44-51. https://www.ncbi.nlm.nih.gov/pubmed/19036639
- ↑ Neafsey DE, Galinsky K, Jiang RH, Young L, Sykes SM, Saif S, Gujja S, Goldberg JM, Young S, Zeng Q, Chapman SB, Dash AP, Anvikar AR, Sutton PL, Birren BW, Escalante AA, Barnwell JW, Carlton JM. 2012. The malaria parasite Plasmodium vivax exhibits greater genetic diversity than Plasmodium falciparum. Nat Genet. 44:1046-50. https://www.ncbi.nlm.nih.gov/pubmed/22863733
- ↑ Arisue N, Kawai S, Hirai M, Palacpac NM, Jia M, Kaneko A, Tanabe K, Horii T. 2011. Clues to Evolution of the SERA Multigene Family in 18 Plasmodium Species. PLoS One. 6: e17775. http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0017775
- ↑ Prasad R, Atul, Soni A, Puri SK, Sijwali PS. 2012. Expression, characterization, and cellular localization of knowpains, papain-like cysteine proteases of the Plasmodium knowlesi malaria parasite. PLoS One. 12:e51619. https://www.ncbi.nlm.nih.gov/pubmed/23251596
- ↑ Brömme D. 2001. Papain-like cysteine proteases. Curr Protoc Protein Sci. 21. doi: 10.1002/0471140864.ps2102s21. https://www.ncbi.nlm.nih.gov/pubmed/18429163
- ↑ Arisue N, Hirai M, Arai M, Matsuoka H, Horii T. 2007. Phylogeny and evolution of the SERA multigene family in the genus Plasmodium. J Mol Evol. 65:82-91. http://link.springer.com/article/10.1007%2Fs00239-006-0253-1
- ↑ Arisue N, Kawai S, Hirai M, Palacpac NM, Jia M, Kaneko A, Tanabe K, Horii T. 2011. Clues to Evolution of the SERA Multigene Family in 18 Plasmodium Species. PLoS One. 6: e17775. http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0017775
- ↑ World Health Organization. (2015). World Malaria Report 2015. Retrieved from http://www.who.int/malaria/publications/world-malaria-report-2015/report/en/
- ↑ Garamszegi LZ. 2009. Patterns of co-speciation and host switching in primate malaria parasites. Malar J. 110. doi: 10.1186/1475-2875-8-110. https://www.ncbi.nlm.nih.gov/pubmed/19463162
- ↑ Ta TH, Hisam S, Lanza M, Jiram AI, Ismail N, Rubio JM. 2014. First case of a naturally acquired human infection with Plasmodium cynomolgi. Malar J. 13: 68. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3937822/
- ↑ Singh B, Daneshvar C. 2013. Human infections and detection of Plasmodium knowlesi. Clin Microbiol Rev. 26:165-84. https://www.ncbi.nlm.nih.gov/pubmed/23554413
- ↑ Prugnolle F, Durand P, Neel C, Ollomo B, Ayala FJ, Arnathau C, Etienne L, Mpoudi-Ngole E, Nkoghe D, Leroy E, Delaporte E, Peeters M, Renaud F. 2010. African great apes are natural hosts of multiple related malaria species, including Plasmodium falciparum. Proc Natl Acad Sci U S A. 107:1458-63. https://www.ncbi.nlm.nih.gov/pubmed/20133889
- ↑ Duval L, Fourment M, Nerrienet E, Rousset D, Sadeuh SA, Goodman SM, Andriaholinirina NV, Randrianarivelojosia M, Paul RE, Robert V, Ayala FJ, Ariey F. 2010. African apes as reservoirs of Plasmodium falciparum and the origin and diversification of the Laverania subgenus. Proc Natl Acad Sci U S A. 107:10561-6. https://www.ncbi.nlm.nih.gov/pubmed/20498054
- ↑ Krizanauskiene A, Hellgren O, Kosarev V, Sokolov L, Bensch S, Valkiunas G. 2006. Variation in host specificity between species of avian haemosporidian parasites: evidence from parasite morphology and cytochrome B gene sequences. J Parasitol. 6:1319-24. https://www.ncbi.nlm.nih.gov/pubmed/17304814
- ↑ Duval L, Robert V, Csorba G, Hassanin A, Randrianarivelojosia M, Walston J, Nhim T, Goodman SM, Ariey F. 2007. Multiple host-switching of Haemosporidia parasites in bats. Malar J. 6:157. https://www.ncbi.nlm.nih.gov/pubmed/18045505