Syntenic path assembly
Contents
- 1 Overview
- 2 Algorithm
- 3 E. coli
- 4 Tetraodon (puffer fish): Takifugu rubripes and Tetraodon nigroviridis
- 5 Carnivora: Giant Panda (WGS Assembly) to Dog (reference genome)
- 6 Arabidopsis ecotypes: Columbia versus Landsberg erecta
- 7 Phoenix dactylifera L. (date palm) v. Oryza sativa japonica (Rice)
- 8 Cannabis sativa (marijuana) v. Prunus persica (peach)
Overview
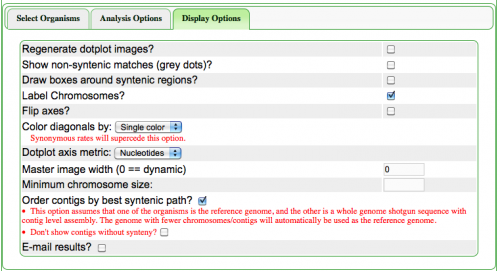
The Syntenic Path Assembly is an option in SynMap to do a reference genome assembly of contigs using synteny to determine the order and orientation of the contigs. To use this option, select "Order contigs by best syntenic path" under the Display Options tab. When an assembly is generated, you may download the Pseudoassembled sequence (contigs are joined using 100 "N"s. "N" is the Ambiguous nucleotide and while it may represent any nucleotide (A, T C, G), this permits the identification of where contigs were "glued" together by this algorithm.).
This algorithm also works quite well in aligning a WGS assembly between distally related organisms. (See below for examples.) Note: Caution needs to be taken about using and trusting a syntenic path assembly. Breakpoints in genome assembly are often due to stretches of repetitive sequence, which can also serve as the sites for genomic rearrangements such as inversions, duplications, and chromosome fusions and fissions.
Algorithm
As implemented in SynMap:
- Parse syntenic blocks from SynMap output file:
- Score (if not present, equal to the number of gene pairs in block)
- Orientation (forward or reverse)
- Names of chromosomes/contigs involved in block
- Start position:
- Remove the first and last gene-pair (often are noisy), if there are more 3 or more gene pairs.
- Calculate the mean start value of all the remaining gene-pairs in a syntenic block
- E.g.: 5 gene pairs with start values of 10, 1000, 1200, 1400, 5000. Start value = (1000+1200+1400)/3 = 1200
- Sum all synteny scores for each pair of chromosomes/contigs between the two genomes
- E.g. if Contig1 and Chr1 have two syntenic blocks, scores 5 and 6, their combined synteny score is 11. (New: May 2012)
- Determine which genome is the reference genome due to having fewer chromosomes/contigs
- For the examples listed here, the reference genome is assumed to have chromosomes, and the genome to be assembled has contigs
- Determine assignment of a contig to a chromosome based on having the highest combined synteny score
- E.g. if Contig1 and Chr1 have a combined synteny score of 11, and Contig1 and Chr2 have a combined synteny store of 8, Contig1 will be assigned to be mapped to Chr1
- Order contigs to a chromsome based on start position (calculation described above)
- Orient contig based on the majority rule of the orientation of the syntenic blocks.
- Specificly, if half or more of the syntenic blocks were in reverse orientation, the contig is reverse complemented.
Thus, the tiling looks like:
|-----A-----| |-----B-----| |-----C-----| |-1-||-2-||-3-| |-4-||-5-||-6-| |-7-||-8-||-9-| etc.,
- Improvements (May 2012)
- Selection of which contig maps to which chromosome has been update to be
- The maximum combined score of all syntenic blocks
- For example. Contig1 has two syntenic blocks with Chr1 (scores 5 and 5); one syntenic block with Chr2 (score 8). Contig1 is placed with Chr1.
- Selection of which contig maps to which chromosome has been update to be
Old Version
- Identify and score each syntenic region
- Assign genome with fewer chromosomes/contigs to be reference
- For each chromosome in reference genome:
- sort all syntenic blocks that match reference genome according to position in reference genome
- for ties in position, pick the syntenic block with the greater synteny score
- flip contig if syntenic block is reversed
Thus, the tiling looks like:
|-----A-----| |-----B-----| |-----C-----| |-1-||-2-||-3-| |-4-||-5-||-6-| |-7-||-8-||-9-| etc.,
Definitions
- WGS: Whole-genome shotgun
E. coli
- Syntenic Path Assembly in SynMap of E. coli
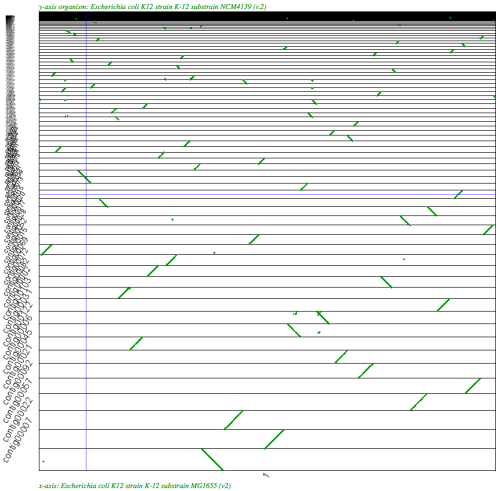
SynMap analysis of a WGS assembly of a strain of E. coli K12 (y-axis) to a reference assembly (x-axis). Results may be regenerated at http://genomevolution.org/r/2k70
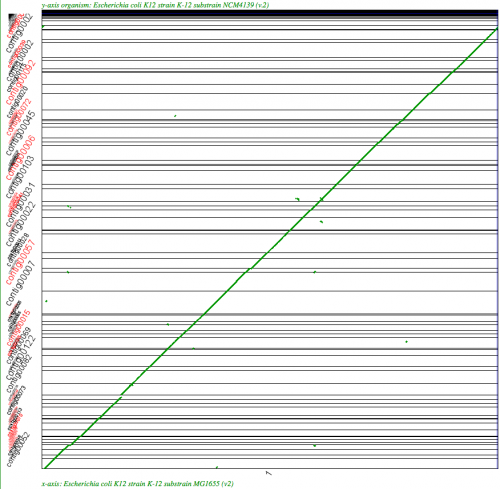
SynMap analysis of a WGS assembly of a strain of E. coli K12 (y-axis) to a reference assembly (x-axis) using SynMap's Syntenic Path Assembly to order contigs. Results may be regenerated at: http://genomevolution.org/r/2k71
Tetraodon (puffer fish): Takifugu rubripes and Tetraodon nigroviridis
Syntenic dotplot using SynMap between two puffer fish, Takifugu rubripes (x-axis) and Tetraodon nigroviridis (y-axis). (Fugu is a WGS assembly.) Note: the background is colored black due to the number of contigs in the WGS assembly. Each contig/chromosome is visually separated from one another by a vertical or horizontal black line. Results may be regenerated at http://genomevolution.org/r/2k7b
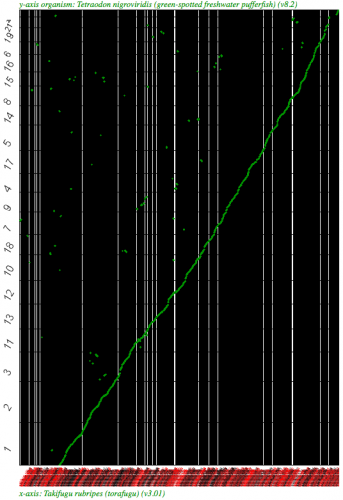
Syntneic dotplot using SynMap between two puffer fish, Takifugu rubripes (x-axis) and Tetraodon nigroviridis (y-axis) using the Syntenic Path Assembly option. Fugu is a WGS assembly and fully syntenic converage to Tetraodon is detectable. Results may be regenerated at http://genomevolution.org/r/2k79 . Note: the background is colored black due to the number of contigs in the WGS assembly. Each contig/chromosome is visually separated from one another by a vertical or horizontal black line.
Carnivora: Giant Panda (WGS Assembly) to Dog (reference genome)
Syntenic dotplot by SynMap of WGS giant panda genome (x-axis) versus complete dog genome(y-axis). Results may be regenerated at http://genomevolution.org/r/2kbv . Note: the background is colored black due to the number of contigs in the WGS assembly. Each contig/chromosome is visually separated from one another by a vertical or horizontal black line.
SynMap Syntneic path assembly of WGS giant panda genome (x-axis) to complete dog genome (y-axis). Results may be regenerated at: http://genomevolution.org/r/2kbt . Note: the background is colored black due to the number of contigs in the WGS assembly. Each contig/chromosome is visually separated from one another by a vertical or horizontal black line.
Arabidopsis ecotypes: Columbia versus Landsberg erecta
Syntenic dotplot by [SynMap]. X-axis Columbia; Y-axis Landsberg. Results may be regenerated: http://genomevolution.org/r/3oke
Syntenic dotplot by [SynMap]. X-axis Columbia; Y-axis Landsberg. Results may be regenerated: http://genomevolution.org/r/3okf
Phoenix dactylifera L. (date palm) v. Oryza sativa japonica (Rice)
Syntenic dotplot by [SynMap]. X-axis Oryza sativa japonica (Rice); Y-axis Phoenix dactylifera L. (date palm). Results may be regenerated: http://genomevolution.org/r/3qjk
Syntenic dotplot by [SynMap]. X-axis Oryza sativa japonica (Rice); Y-axis Phoenix dactylifera L. (date palm). Results may be regenerated: http://genomevolution.org/r/3qjn
Cannabis sativa (marijuana) v. Prunus persica (peach)
Syntenic dotplot by [SynMap]. X-axis Cannabis sativa (marijuana); Prunus persica (peach). Results may be regenerated: http://genomevolution.org/r/3wz5
Syntenic dotplot by [SynMap]. X-axis Cannabis sativa (marijuana); Prunus persica (peach). Results may be regenerated: http://genomevolution.org/r/3wz3