Creosote
From CoGepedia
Contents
Twig2Genome Notes
Assembly
Loading into CoGe
SoapDeNovo assemly: 1,570,116 contigs (370MB): http://genomevolution.org/CoGe/OrganismView.pl?dsgid=12183 (Note: too many contigs to process and visualize in SynMap)
SoapDeNovo assembly of contigs >= 2000nt: 6,976 contigs (20MB): http://genomevolution.org/CoGe/OrganismView.pl?dsgid=12185
ABySS assembly (bpsize=64, 2kb minimum contig size): 122,972 contigs (392MB): http://genomevolution.org/CoGe/OrganismView.pl?dsgid=12275
- Syntenic Path Assembly to peach: http://genomevolution.org/r/3xmq
Velvet assembly 515,190 contigs (241MB): http://genomevolution.org/CoGe/OrganismView.pl?dsgid=12245
CLC4 assembly 685,475 contigs (508MB): http://genomevolution.org/CoGe/OrganismView.pl?dsgid=12244
- Syntenic Path Assembly to peach: http://genomevolution.org/r/3xpd
Syntenic Path Assembly
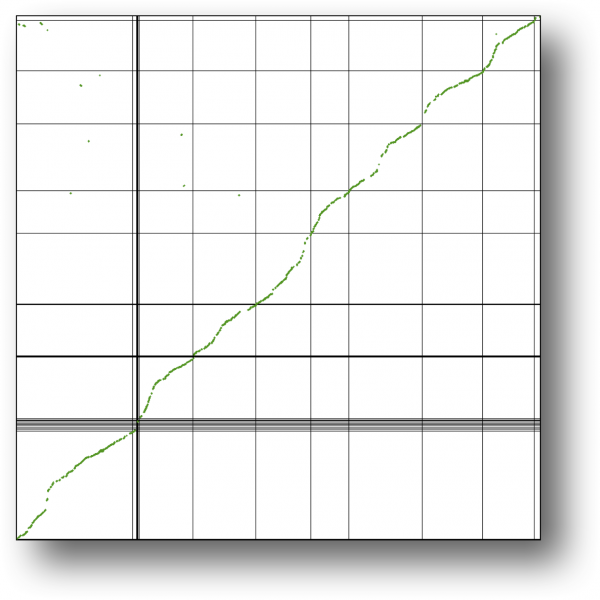
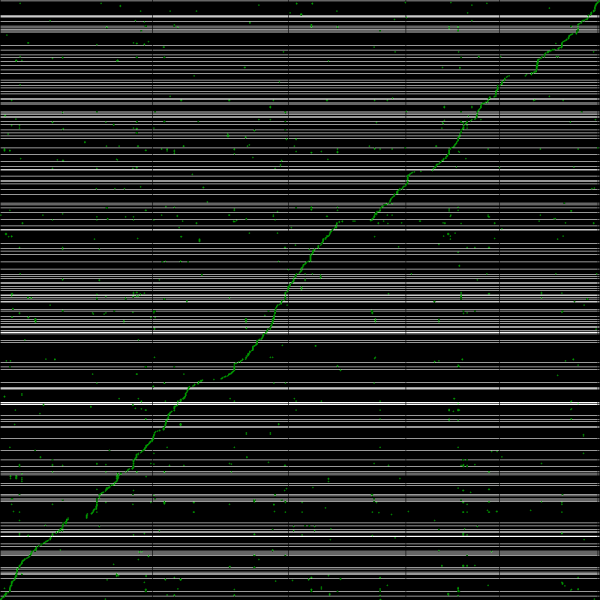
Syntenic path assembly with SynMap of creosote (x-axis) and peach (y-axis). Results may be regenerated at: http://genomevolution.org/r/3w95
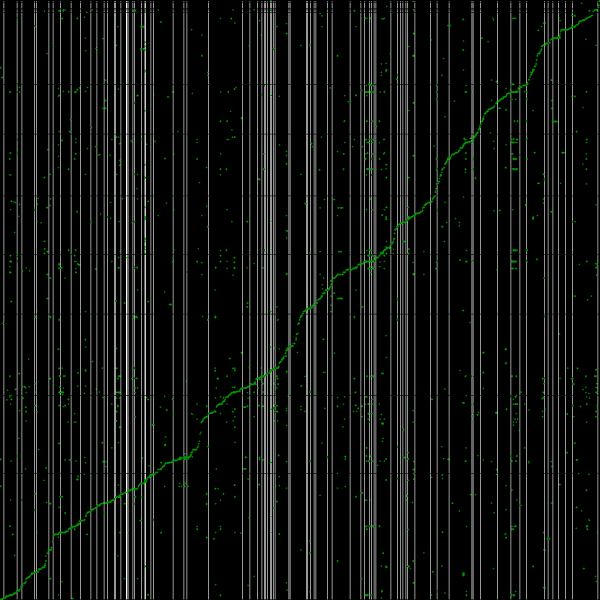
Syntenic path assembly with SynMap of creosote (y-axis) and Arabidopsis thaliana Col-0 (x-axis). Results may be regenerated at: http://genomevolution.org/r/3w96
Pseudo-Assembly
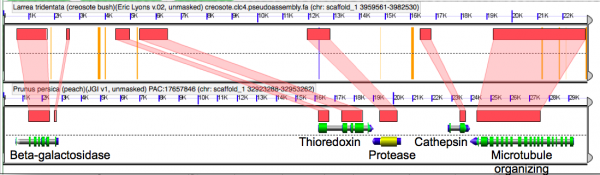
Microsynteny analysis of the pseudo-assembled creosote genome to the peach genome. Orange bars are unsequenced Ns that represent contigs glued together in creosote by the syntenic path assembly method. Note the concordance of gene model coding sequences.