GEvo Blastn Bug
Contents
Bug Description
When using blastn in GEvo, a large HSP appear in the middle of the region when the query sequence length is changed by 1 nucleotide.
This problem was identified by Mike Freeling.
Visualization
Difference between analyses is that addition of 1 nucleotide to the top panel of the top analysis

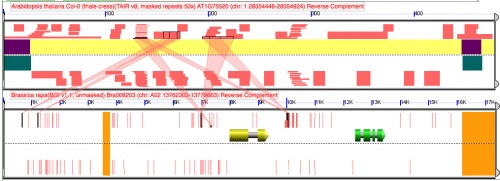
Report of "disappearing" HSP in blast file
Top portion of blast report which contains the large HSP (problem HSP starts at "Query 129"):
BLASTN 2.2.24+
Query= AT1G75520
Length=478
Subject= Bra008203
Length=17361
Score = 68.0 bits (35), Expect = 3e-14
Identities = 76/94 (80%), Gaps = 4/94 (4%)
Strand=Plus/Plus
Query 1 CTAGGTTTCGTGTTCCACTGATCAAAGATTTGAAAAAAAACATATACTTAGTAAACTTCA 60
|||||||| ||||||||||||||||||| || || ||||||| | |||||| |
Sbjct 9943 CTAGGTTTTGTGTTCCACTGATCAAAGAGTT----AAGAACATATTTCAATAAAACTTTA 9998
Query 61 AGCAATTTTTATATTACCCAATTGAATTTCTCCA 94
| |||||||||| |||||| |||||||||||||
Sbjct 9999 AATAATTTTTATACTACCCAGTTGAATTTCTCCA 10032
Score = 54.5 bits (28), Expect = 3e-10
Identities = 32/34 (94%), Gaps = 0/34 (0%)
Strand=Plus/Plus
Query 442 TTGGCGTAGATGAATGTAAACGGATGGTAATATA 475
|||||||| |||||||||||||||| ||||||||
Sbjct 10342 TTGGCGTATATGAATGTAAACGGATAGTAATATA 10375
Score = 50.7 bits (26), Expect = 4e-09
Identities = 77/95 (81%), Gaps = 5/95 (5%)
Strand=Plus/Plus
Query 129 ATATATATATACCCAACAACTGAGAAAAGATGGAAAAAGTTTAGTTAAAAACTGGTCCTG 188
||||||||||| |||||| || |||||||||||| || || ||||||||| | |
Sbjct 10072 ATATATATATAACCAACATCTCGGAAAAGATGGAACAAATT-AGTTAAAAAAA---CATT 10127
Query 189 GGCGGCTTTAAATTATATTTATGCACTTAAATTTA 223
||||||||||||| ||||| ||| | |||||||||
Sbjct 10128 GGCGGCTTTAAATCATATTCATGTA-TTAAATTTA 10161
Score = 48.8 bits (25), Expect = 2e-08
Identities = 47/58 (81%), Gaps = 0/58 (0%)
Strand=Plus/Plus
Query 316 GGTTTTTACTTAGATAATATCGTGTCATTCCATCTAGATTCAACCCCTGTCTACAATA 373
|||||| ||| |||| |||| || || | | ||||||||||||||| |||||||||
Sbjct 10207 GGTTTTCACTGAGATGATATTGTTCCAGTTCCACTAGATTCAACCCCTCTCTACAATA 10264
Score = 25.7 bits (13), Expect = 0.15
Identities = 13/13 (100%), Gaps = 0/13 (0%)
Strand=Plus/Plus
Query 127 TAATATATATATA 139
|||||||||||||
Sbjct 7327 TAATATATATATA 7339
Top portion of blast report which DOES NOT contains the large HSP :
BLASTN 2.2.24+
Query= AT1G75520
Length=479
Subject= Bra008203
Length=17361
Score = 69.9 bits (36), Expect = 7e-15
Identities = 77/95 (81%), Gaps = 4/95 (4%)
Strand=Plus/Plus
Query 1 ACTAGGTTTCGTGTTCCACTGATCAAAGATTTGAAAAAAAACATATACTTAGTAAACTTC 60
||||||||| ||||||||||||||||||| || || ||||||| | ||||||
Sbjct 9942 ACTAGGTTTTGTGTTCCACTGATCAAAGAGTT----AAGAACATATTTCAATAAAACTTT 9997
Query 61 AAGCAATTTTTATATTACCCAATTGAATTTCTCCA 95
|| |||||||||| |||||| |||||||||||||
Sbjct 9998 AAATAATTTTTATACTACCCAGTTGAATTTCTCCA 10032
Score = 54.5 bits (28), Expect = 3e-10
Identities = 32/34 (94%), Gaps = 0/34 (0%)
Strand=Plus/Plus
Query 443 TTGGCGTAGATGAATGTAAACGGATGGTAATATA 476
|||||||| |||||||||||||||| ||||||||
Sbjct 10342 TTGGCGTATATGAATGTAAACGGATAGTAATATA 10375
Score = 48.8 bits (25), Expect = 2e-08
Identities = 47/58 (81%), Gaps = 0/58 (0%)
Strand=Plus/Plus
Query 317 GGTTTTTACTTAGATAATATCGTGTCATTCCATCTAGATTCAACCCCTGTCTACAATA 374
|||||| ||| |||| |||| || || | | ||||||||||||||| |||||||||
Sbjct 10207 GGTTTTCACTGAGATGATATTGTTCCAGTTCCACTAGATTCAACCCCTCTCTACAATA 10264
Score = 25.7 bits (13), Expect = 0.15
Identities = 13/13 (100%), Gaps = 0/13 (0%)
Strand=Plus/Plus
Query 128 TAATATATATATA 140
|||||||||||||
Sbjct 7327 TAATATATATATA 7339
Top portion of blast report which contains the large HSP :br>
Lambda K H
1.33 0.621 1.12
Gapped
Lambda K H
1.33 0.621 1.12
Effective search space used: 8066820
Matrix: blastn matrix 1 -2
Gap Penalties: Existence: 5, Extension: 2
Top portion of blast report which DOES NOT contains the large HSP :
Lambda K H
1.33 0.621 1.12
Gapped
Lambda K H
1.33 0.621 1.12
Effective search space used: 8084168
Matrix: blastn matrix 1 -2
Gap Penalties: Existence: 5, Extension: 2
Blast Commands
Extra HSP, Legacy Blast:
/usr/local/bin/legacy_blast.pl bl2seq -p blastn -o /opt/apache/CoGe/tmp/GEvo/52376346_1-2.bl2seq -i /opt/apache/CoGe/tmp/GEvo/f34524790ds39598r1c1u-2233d777g2dsg3.faa -j /opt/apache/CoGe/tmp/GEvo/f103007203ds48732r1cA02u8000d8000g1dsg12468.faa -W 7 -G 5 -E 2 -q -2 -r 1 -e 30 -F F
Extra HSP, Blast+
blastn -query /opt/apache/CoGe/tmp/GEvo/f34524790ds39598r1c1u-2233d777g2dsg3.faa -subject /opt/apache/CoGe/tmp/GEvo/f103007203ds48732r1cA02u8000d8000g1dsg12468.faa -evalue 30 -gapopen 5 -gapextend 2 -word_size 7 -penalty -2 -reward 1 -dust no
Missing HSP, Legacy Blast
/usr/local/bin/legacy_blast.pl bl2seq -p blastn -o /opt/apache/CoGe/tmp/GEvo/22224470_1-2.bl2seq -i /opt/apache/CoGe/tmp/GEvo/f34524790ds39598r1c1u-2232d777g2dsg3.faa -j /opt/apache/CoGe/tmp/GEvo/f103007203ds48732r1cA02u8000d8000g1dsg12468.faa -W 7 -G 5 -E 2 -q -2 -r 1 -e 30 -F F
Missing HSP, Blast+
blastn -query /opt/apache/CoGe/tmp/GEvo/f34524790ds39598r1c1u-2232d777g2dsg3.faa -subject /opt/apache/CoGe/tmp/GEvo/f103007203ds48732r1cA02u8000d8000g1dsg12468.faa -evalue 30 -gapopen 5 -gapextend 2 -word_size 7 -penalty -2 -reward 1 -dust no
Conclusions
- The bug is in blastn. Current version is 2.2.24
- Possible causes:
- Change in sequence search space (sequence length) causes change in evalue
- Change in sequence causes an "edge-effect" where exact sequence pattern at the end of the character sequence causes a chance in HSP identification
- Repeat sequences are causing an internal problem to how blast identifies and categorizes HSPs
Download
The sequences, blast reports, log files, and GEvo Images can be obtained at: http://genomevolution.org/CoGe/data/distrib/gevo_blast_bug.tar.gz
Comparison to Blast 2.2.25+
Same problem exists: GEvo bug Blast 2.2.25+