Plasmodium analysis workflow 3: Tools useful on the study of multigene families
Contents
Outline
RETURN TO THE MAIN PAGE: Using_CoGe_for_the_analysis_of_Plasmodium_spp
RETURN TO THE FIRST WORKFLOW: Plasmodium analysis workflow 1: Tools that evaluate genomic properties and amino acid usage
RETURN TO THE PREVIOUS WORKFLOW: Plasmodium analysis workflow 2: Tools for the syntenic analysis of whole genomes and microsyntenic regions
Abstract
Gene duplication has long been recognized as an important source for genome innovation and adaptation. In the study of multigene families’ evolution, the identification of evolutionary relations, e.g. paralogy and orthology, between family members is fundamental. Plasmodium spp. multigene families perform a wide array of functions; have diverse gene organization, and distinct evolutionary histories. Here, we show CoGe tools that can be used to evaluate evolutionary patterns of Plasmodium-specific and species-specific multigene families.
Identifying gene homologs (CoGeBLAST)
The identification of homology based on sequence similarity is a key tool for gaining insight into an organism’s biology and genetics. Defining evolutionary relationships and inferring common ancestry is particularly challenging when dealing with multigene families. Plasmodium multigene families perform a wide array of functions, have diverse gene organization, and distinct evolutionary histories. Here we focus on a set of multi-gene families arising from the subtelomere (e.g. var, stevor, rifin, or vir) that have very complex evolutionary patterns and organizations [1]. These four gene families are of particular interest because of their role in immune evasion and cell invasion. In addition, these families have undergone rapid sequence evolution and gene turnover [2][3][4]. These factors make inferring orthology/paralogy and gene gain/loss events in Plasmodium subtelomeric families a complex task.
The 313 members of P. vivax’s vir family are grouped into 10 subfamilies based on their sequence similarity. Gene size and structure (number of exons) is largely variable among family members [5][6][7]. The genetic diversity in the vir family is larger than that of other P. vivax families. Only fifteen of the 313 vir genes are shared across all sequenced P. vivax strains despite the recent emergence of the species ~ five million years ago. Within this group, PVX_113230 has been proposed as a potential family founder based on its high sequence conservation [8].
Here we use CoGeBLAST to identify the proposed founder of the Plasmodium vir family (PVX_113230) in six P. vivax strains (including the recently sequenced PO1 strain). CoGeBLAST incorporates genome visualization into BLAST analyses. Therefore, this tool facilitates the study of complex evolutionary patterns.

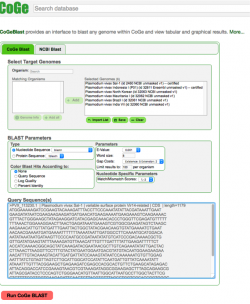
| The following steps show how to use CoGeBLAST in the CoGe platform:
2. Click on CoGeBLAST or follow this link: https://genomevolution.org/coge/CoGeBlast.pl 3. Type the scientific name of the Organism of interest in the Search box. All genomes with names matching the search term will appear under the Matching Organisms menu. Notebooks matching the term will appear in a new window after clicking on Import List. 4. Select all the genomes of interest and click on + Add. The genomes will now appear on the Selected Genomes menu. You can also select any of your Notebooks and include all the genomes contained in it. 5. Enter your query sequence in FASTA format. If desired, you can change the BLAST Parameters before starting the analysis. 6. Once all information is included click on Run CoGe BLAST (Figure 1). 7. The analysis output will include:
You can find links to the FASTA sequences used in this analysis in the "Sample data" section at the end of this page. |
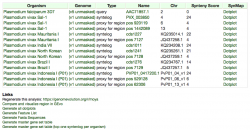
Sequences with significant similarity to PVX_113230 were found in all the evaluated P. vivax strains, including PO1. However, the number of high-scoring segment pairs for each P. vivax genome was variable. The highest number of sequence homologs was observed in the strains: Mauritania, PO1, and Salvador-1. Sequence divergence of vir members within P. vivax seems to affect the number of high-scoring segment pairs per strain. Thus, the variation in the number of HSPs across strains further supports observations about the high sequence variation among vir homologs.
The location of HSPs appears to be slightly variable across genomes. However, we cannot confirm this patterns until the Mauritania, North Korea, Brazil I, and India VII genomes are fully assembled. Between the two fully assembled P. vivax genomes (Salvador-1 and PO1), BLAST hits were located largely in the same chromosome regions (Figure 2). As expected, a higher number of BLAST hits and a more variable genome location were observed when a less conserved vir family member (PVX_096004.1) was used as a query (analysis can be run following this link: https://genomevolution.org/r/mkcg).
Identifying sets of syntenic genes amongst several genomes (SynFind)
Small-scale genomic rearrangements are often linked to species-specific gene gain/loss events. Family-linked rearrangements are observed amongst closely related Plasmodium species, and in occasion, at an intra-specific level. CoGe’s tool, SynFind, is used to identify gene homologs across any number of genomes, and thus can be of use to identify these rearrangements.
The evolutionary trajectory of multigene families can be difficult to infer, especially in those with a scattered organization or rapid gene turnover. While this issue is particularly prevalent in species-specific gene families; genus-specific families can present intricate evolutionary patterns as well. One good example can be found in the SERA (serine repeat antigen) family, a gene family that has experienced a significant number of inter-specific contractions, expansions, and rearrangements. These patterns remain to be evaluated at an intra-specific level. We will use SynFind to study family's organization of SERA paralogs in 6 P. vivax strains.
SERA paralogs are expressed during various stages of the Plasmodium life cycle. All SERA family members encode proteins with a papain-like cysteine protease motif [9]. These motifs are commonly found both inside and outside the genus Plasmodium [10][11]. One member (SERA-5), expressed during late trophozoite and schizont stages, has been considered as a promising malaria vaccine target [12]. We will use this gene sequence as a query for the SynFind analysis.
| The following steps show how to use SynFind:
2. Click on SynFind or follow this link: https://genomevolution.org/CoGe/SynFind.pl. 3. Type a scientific name of your search bar under Select Target Genomes. Organisms and genomes with names matching the search term will be displayed on the Matching Organisms menu. 4. Select the genomes of interest using Ctl+click or Command+click, then click on + Add. The genomes will appear on the Selected Genomes menu. You can also import genomes from your Notebooks. 5. Type the Name, Annotation, or Organisms on the Specify Features section. It is recommended to include as many specific terms as possible. Once done click on Search. 6. All matches to the search term and the genome where they have been found will appear in a new menu within the same section. Select all relevant Matches and the reference Genome. 7. Click on Run SynFind to start the analysis. 8. SynFind will output all syntenic regions from the reference genome and their Syntenic depth. This output can be used as a query for other CoGe tools.
GEvo results can be replicated here: https://genomevolution.org/r/mpdf |
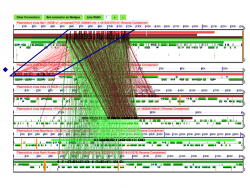
We used Synfind to identify genes homologous to SERA-5 across 6 P. vivax genomes (Figure 3). Synfind’s output was used as a query for a GEvo analysis of the region. Our results show a conserved number of SERA paralogs in all P. vivax strains. The organization of the SERA family was different on the Brazil I strain respect to other P. vivax strains (Figure 4). Previous studies on SERA have suggested that some family members are unique to P. vivax and closely related species [13]. Our results indicate that family organization is not completely conserved on the intra-specific level. This is most evident on recently duplicated paralogs.
SynFind also identified matching segments outside the SERA multigene family. These segments belonged to hypothetical protein coding genes, ATP proteases, and uncharacterized transcripts. Papain-like cysteine protease motifs are commonly found outside both Plasmodium and the SERA family. Thus, is likely that these segments share a papain-like cysteine protease motif but are not evolutionarily related to SERA.
Outline
RETURN TO THE MAIN PAGE: Using_CoGe_for_the_analysis_of_Plasmodium_spp
RETURN TO THE FIRST WORKFLOW: Plasmodium analysis workflow 1: Tools that evaluate genomic properties and amino acid usage
RETURN TO THE PREVIOUS WORKFLOW: Plasmodium analysis workflow 2: Tools for the syntenic analysis of whole genomes and microsyntenic regions
References
- ↑ Singh V, Gupta P, Pande V. 2014. Revisiting the multigene families: Plasmodium var and vir genes. J Vector Borne Dis. 51:75-81. https://www.ncbi.nlm.nih.gov/pubmed/24947212
- ↑ Niang M, Yan Yam X, Preiser PR. 2009. The Plasmodium falciparum STEVOR multigene family mediates antigenic variation of the infected erythrocyte. PLoS Pathog. 5:e1000307. https://www.ncbi.nlm.nih.gov/pubmed/19229319
- ↑ Witmer K, Schmid CD, Brancucci NM, Luah YH, Preiser PR, Bozdech Z, Voss TS. 2012. Analysis of subtelomeric virulence gene families in Plasmodium falciparum by comparative transcriptional profiling. Mol Microbiol. 84:243-59. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3491689/
- ↑ Petter M, Bonow I, Klinkert MQ. 2008. Diverse expression patterns of subgroups of the rif multigene family during Plasmodium falciparum gametocytogenesis. PLoS One. 3:e3779. http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0003779
- ↑ Carlton JM, Adams JH, Silva JC, Bidwell SL, Lorenzi H, Caler E, Crabtree J, Angiuoli SV, Merino EF, Amedeo P, Cheng Q, Coulson RM, Crabb BS, Del Portillo HA, Essien K, Feldblyum TV, Fernandez-Becerra C, Gilson PR, Gueye AH, Guo X, Kang'a S, Kooij TW, Korsinczky M, Meyer EV, Nene V, Paulsen I, White O, Ralph SA, Ren Q, Sargeant TJ, Salzberg SL, Stoeckert CJ, Sullivan SA, Yamamoto MM, Hoffman SL, Wortman JR, Gardner MJ, Galinski MR, Barnwell JW, Fraser-Liggett CM. 2008. Comparative genomics of the neglected human malaria parasite Plasmodium vivax. Nature. 455:757-63. https://www.ncbi.nlm.nih.gov/pubmed/18843361
- ↑ Lopez FJ, Bernabeu M, Fernandez-Becerra C, del Portillo HA. 2013. A new computational approach redefines the subtelomeric vir superfamily of Plasmodium vivax. BMC Genomics. 14:8. https://www.ncbi.nlm.nih.gov/pubmed/?term=A+new+computational+approach+redefines+the+subtelomeric+vir+superfamily+of+Plasmodium+vivax
- ↑ Fernandez-Becerra C, Yamamoto MM, Vêncio RZ, Lacerda M, Rosanas-Urgell A, del Portillo HA. 2009. Plasmodium vivax and the importance of the subtelomeric multigene vir superfamily. Trends Parasitol. 2009 25:44-51. https://www.ncbi.nlm.nih.gov/pubmed/19036639
- ↑ Neafsey DE, Galinsky K, Jiang RH, Young L, Sykes SM, Saif S, Gujja S, Goldberg JM, Young S, Zeng Q, Chapman SB, Dash AP, Anvikar AR, Sutton PL, Birren BW, Escalante AA, Barnwell JW, Carlton JM. 2012. The malaria parasite Plasmodium vivax exhibits greater genetic diversity than Plasmodium falciparum. Nat Genet. 44:1046-50. https://www.ncbi.nlm.nih.gov/pubmed/22863733
- ↑ Arisue N, Kawai S, Hirai M, Palacpac NM, Jia M, Kaneko A, Tanabe K, Horii T. 2011. Clues to Evolution of the SERA Multigene Family in 18 Plasmodium Species. PLoS One. 6: e17775. http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0017775
- ↑ Prasad R, Atul, Soni A, Puri SK, Sijwali PS. 2012. Expression, characterization, and cellular localization of knowpains, papain-like cysteine proteases of the Plasmodium knowlesi malaria parasite. PLoS One. 12:e51619. https://www.ncbi.nlm.nih.gov/pubmed/23251596
- ↑ Brömme D. 2001. Papain-like cysteine proteases. Curr Protoc Protein Sci. 21. doi: 10.1002/0471140864.ps2102s21. https://www.ncbi.nlm.nih.gov/pubmed/18429163
- ↑ Arisue N, Hirai M, Arai M, Matsuoka H, Horii T. 2007. Phylogeny and evolution of the SERA multigene family in the genus Plasmodium. J Mol Evol. 65:82-91. http://link.springer.com/article/10.1007%2Fs00239-006-0253-1
- ↑ Arisue N, Kawai S, Hirai M, Palacpac NM, Jia M, Kaneko A, Tanabe K, Horii T. 2011. Clues to Evolution of the SERA Multigene Family in 18 Plasmodium Species. PLoS One. 6: e17775. http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0017775