Plasmodium genome analysis using Syntenic Path Assembly
Using Syntenic Path Assembly (SPA) to make analysis of poor or early genome assemblies easier (SynMap - SPA tool)
There are a large number of Plasmodium genomes that remain to be fully sequenced, assembled and annotated. Incomplete genomic data comes from a variety of sources:
- Genomic information published on early assembly stages.
- Partially sequenced genomes.
- Low-quality genome segments.
Sequencing projects can be simplified by the use of a reference genome as a guide for genome assembly. While unassembled and non-annotated genomes can be used in smaller-scale studies (e.g. orthologs can be identified with BLAST), there are limitations in their usability in large-scale comparative genomics.
Tools that generate preliminary assemblies are incredibly important for comparative analyses, especially as more genomic data become available. CoGe’s tool, Syntenic_path_assembly (SPA), creates a graphical display of syntenic gene pairs based on a reference genome. We will use SPA to assemble the P. inui genome (on scaffold level as in 2016) using the fully assembled P. coatneyi genome as a reference.
| The following steps show how to use SynMap - SPA tool:
2. Run SynMap between an assembled and a non-assembled genome (this might take longer than analyses using two fully assembled genomes). 3. After running SynMap click on the Display Options tab and find the SPA tool (Figure 1). Select the tool by clicking on the check mark next to: The Syntenic Path Assembly (SPA)? 4. After a few minutes, the incomplete genome will be assembled using the second genome as a reference.
|
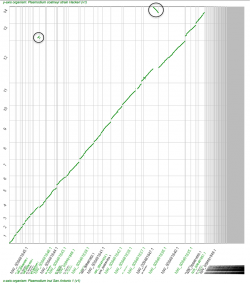
SPA is extremely useful to generate quick and dirty genome assemblies; however, there are some limitations regarding assembly interpretation. We highlight two scenarios seen in the P. inui’s SPA using P. coatneyi’s genome as a reference (Figure 2).
Rearrangement events such as inversions or duplications cannot be identified using SPA. For one, several contigs can be syntenic to the same region of the reference genome without signaling a duplication event. Also, contigs syntenic to a reverse DNA strand might not reflect chromosome inversions (black circles, Figure 2).
In addition, contigs will be arranged to increase synteny between the unassembled and the reference genome. Thus, using different reference genomes will result in different preliminary assemblies. In the case of P. inui, using P. coatneyi (a closely related species) or P. falciparum (a more distantly related species) as reference genomes will result in different assemblies. Therefore, before running SPA, the reference genomes should be selected after consideration of the biological and evolutionary relationship between species. Also, interpretation of SPA assemblies might be problematic when working with transposon-rich genomes.