Difference between revisions of "GEvo Blastn Bug"
From CoGepedia
| Line 7: | Line 7: | ||
Difference between analyses is that addition of 1 nucleotide to the top panel of the top analysis [[Image:Screen Shot 2012-02-11 at 4.55.25 AM.png|thumb|center|500px]] [[Image:Screen Shot 2012-02-11 at 4.57.06 AM.png|thumb|center|500px]] | Difference between analyses is that addition of 1 nucleotide to the top panel of the top analysis [[Image:Screen Shot 2012-02-11 at 4.55.25 AM.png|thumb|center|500px]] [[Image:Screen Shot 2012-02-11 at 4.57.06 AM.png|thumb|center|500px]] | ||
| − | == Report of "disappearing" HSP in blast file == | + | == Report of "disappearing" HSP in blast file == |
| − | Top portion of blast report which contains the large HSP:<br> | + | Top portion of blast report which contains the large HSP (problem HSP starts at "Query 129"):<br> |
<pre>BLASTN 2.2.24+ | <pre>BLASTN 2.2.24+ | ||
Revision as of 06:01, 11 February 2012
Bug Description
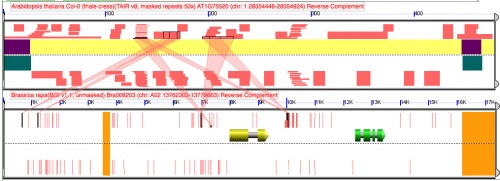
When using blastn in GEvo, a large HSP appear in the middle of the region when the query sequence length is changed by 1 nucleotide.
Visualization
Difference between analyses is that addition of 1 nucleotide to the top panel of the top analysisReport of "disappearing" HSP in blast file
Top portion of blast report which contains the large HSP (problem HSP starts at "Query 129"):
BLASTN 2.2.24+
Query= AT1G75520
Length=478
Subject= Bra008203
Length=17361
Score = 68.0 bits (35), Expect = 3e-14
Identities = 76/94 (80%), Gaps = 4/94 (4%)
Strand=Plus/Plus
Query 1 CTAGGTTTCGTGTTCCACTGATCAAAGATTTGAAAAAAAACATATACTTAGTAAACTTCA 60
|||||||| ||||||||||||||||||| || || ||||||| | |||||| |
Sbjct 9943 CTAGGTTTTGTGTTCCACTGATCAAAGAGTT----AAGAACATATTTCAATAAAACTTTA 9998
Query 61 AGCAATTTTTATATTACCCAATTGAATTTCTCCA 94
| |||||||||| |||||| |||||||||||||
Sbjct 9999 AATAATTTTTATACTACCCAGTTGAATTTCTCCA 10032
Score = 54.5 bits (28), Expect = 3e-10
Identities = 32/34 (94%), Gaps = 0/34 (0%)
Strand=Plus/Plus
Query 442 TTGGCGTAGATGAATGTAAACGGATGGTAATATA 475
|||||||| |||||||||||||||| ||||||||
Sbjct 10342 TTGGCGTATATGAATGTAAACGGATAGTAATATA 10375
Score = 50.7 bits (26), Expect = 4e-09
Identities = 77/95 (81%), Gaps = 5/95 (5%)
Strand=Plus/Plus
Query 129 ATATATATATACCCAACAACTGAGAAAAGATGGAAAAAGTTTAGTTAAAAACTGGTCCTG 188
||||||||||| |||||| || |||||||||||| || || ||||||||| | |
Sbjct 10072 ATATATATATAACCAACATCTCGGAAAAGATGGAACAAATT-AGTTAAAAAAA---CATT 10127
Query 189 GGCGGCTTTAAATTATATTTATGCACTTAAATTTA 223
||||||||||||| ||||| ||| | |||||||||
Sbjct 10128 GGCGGCTTTAAATCATATTCATGTA-TTAAATTTA 10161
Score = 48.8 bits (25), Expect = 2e-08
Identities = 47/58 (81%), Gaps = 0/58 (0%)
Strand=Plus/Plus
Query 316 GGTTTTTACTTAGATAATATCGTGTCATTCCATCTAGATTCAACCCCTGTCTACAATA 373
|||||| ||| |||| |||| || || | | ||||||||||||||| |||||||||
Sbjct 10207 GGTTTTCACTGAGATGATATTGTTCCAGTTCCACTAGATTCAACCCCTCTCTACAATA 10264
Score = 25.7 bits (13), Expect = 0.15
Identities = 13/13 (100%), Gaps = 0/13 (0%)
Strand=Plus/Plus
Query 127 TAATATATATATA 139
|||||||||||||
Sbjct 7327 TAATATATATATA 7339