CoGe system architecture
From CoGepedia

CoGe’s system architecture. At CoGe’s core is a relational database (a) designed to store multiple versions of genomes from multiple organisms in any state of assembly. On top of this database is an Application Programming Interface (API; 2) that provides high-level functional access to the database. Using this API, several genome loading programs (3) scour various genomic repositories to add new genomes to the database. CoGe’s utilizes several new libraries for visualization genomic data at a variety of scales (4) and many third party applications for sequence analysis (5). A suite of web-based applications (6) tie together these subsystems to create an interconnected set of software tools assessable to researchers anywhere in the world.
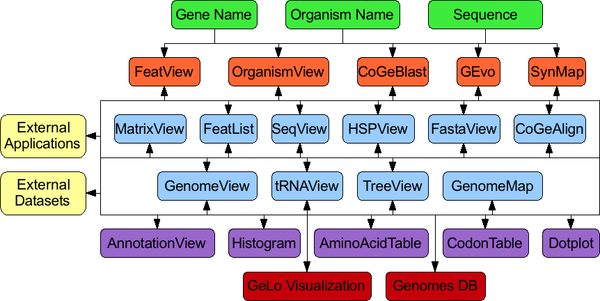
CoGe’s suite of web-based applications creates an open-ended analysis network. The interconnected nature of these tools (black lines and arrows) create an open-ended analysis network with no pre-defined beginning or end of an analysis. Each tool is specialized for a particular type of data or analysis, has its own web-application. Using a web-based framework for tool development inherently creates an method for the storage of analysis states so an analysis can be “saved” for future work. This also allows external tools and datasets to easily link into CoGe for many types of data and analyses. Green boxes are data types that researchers can use to begin an analysis. Orange boxes are applications that allow for direct user access. Blue boxes are internal applications that are accessed through another tool. Purple boxes are modules for web-tools for displaying specific types of data. Red boxes are core modules of CoGe. Yellow boxes are for resources outside of CoGe’s web-based framework.
There are several base components to CoGe:
- Genomes Database for storing any number of genomes from any number of organisms in any state of assembly
- Database API for retrieving data from the database
- Genome Loaders for loading new genomes
- Genomic Visualization Subsystem for visualizing genomic information
- 3rd party applications for doing a lot of other stuff. These are mostly sequence comparison algorithms.
- Web Applications for making genomic data and analytical tools easily accessible.
- CoGe's interconnected tools create open-ended analysis networks.