Difference between revisions of "Analysis of variations found in genomes of Escherichia coli strain K12 DH10B and strain B REL606 using SynMap and GEvo analysis"
| (37 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| − | + | ==Background== | |
| − | + | In this exercise you will compare the genomes of two ''Escherichia coli'' strains, K12 DH10B and B REL606, using whole genome syntenic comparison and high-resolution analyses of specific genomic regions. These analyses will use CoGe's tools [[SynMap]] and [[GEvo]] respectively, and will reveal evolutionary changes between these two genomes that happened after the divergence of their lineages. While the nucleotide sequence of these genomes is identical over large expanses of their genomes, many other types of large-scale genomic change will be discovered including phage insertions, transposon transposition, and genomic insertion, deletion, inversion, and duplication events. The computational tools used to do these analyses can be used for comparing the genomes of any organisms. To learn what organisms and genomes are available in CoGe, please see [[GenomeView]]. | |
| − | [[ | + | ==Generating a [[syntenic dotplot]] of two Escherichia coli strains== |
| + | [[Synteny]], in genomic terms, is defined as two or more genomic regions that are derived from a common ancestor. To identify [[syntenic regions]], you are going generate a [[syntenic dotplot]] of the genomes of ''Echerichia coli'' strains K12 DH10B and B REL606 using [[SynMap]]. First, go to [http://www.synteny.cnr.berkeley.edu/CoGe/Synmap.pl SynMap]. Search for these ''E. coli'' strains in CoGe's database by typing in part of the their names in the "Name" search boxes for Organism 1 and Organism 2. For example, search for "DH10B" and "REL606" respectively, or type "escheri" in both boxes. Alternatively, use [http://synteny.cnr.berkeley.edu/CoGe/SynMap.pl?dsgid1=7454;dsgid2=4241;D=20;g=10;A=5;w=0;b=1;ft1=1;ft2=1;dt=geneorder this link] to load SynMap with these genomes already specified. Once [[SynMap]] has found organisms that matches these names, make sure they are selected in the Organism List. While there are several parameters that can be configured when generating a syntenic dotplot using SynMap, the default settings work well for most situations, and very well for closely related organisms. Click "Generate SynMap" to start the analysis. | ||
| − | + | [[Image:Generate synmap.png|thumb|center|700px| Webpage of SynMap]] | |
| − | + | A lot of processing is happening behind the scenes, but the general way a syntenic dotplot is created is: | |
| + | #All protein coding regions ([[CDS]])) are extracted from each genome | ||
| + | #These sequences are [[blasted]] against each other to identify putative homologous gene pairs | ||
| + | #Putative homologous gene pairs are analyzed to determine if they share a collinear order between the genoems | ||
| + | The general principle is that the most likely and parsimonious way two genomes have a collinear series of homologous genes is those genomic regions in each organism are derived from a common ancestral genomic region (hence they are syntenic). In other words, genomic synteny is inferred by a collinear arrangement of putatively homologous genes in two or more genomic genomic regions. | ||
| − | + | ==Syntenic dotplot data interpretation and analysis== | |
| − | If you look closely at the syntenic dotplot of these genomes, you'll notice that the syntenic line is not perfect, and there are many "breaks" or discontinuities between them. In the figure above, these are indicated by a numbered arrow. These breaks in the syntenic path between these genomes are due to genomic changes happening at a larger scale than a single nucleotide polymorphism, and are mostly likely due to the insertion or deletion of a many nucleotide chunk in one of these genomes. In order to accurately account and characterize these | + | When finished, SynMap will display a dotplot. Each axis of the dotplot is in nucleotide units and represents one of the two genomes laid end to end. The lower-left corner represents the start of each genome (usually 'ORI' for circular bacterial genomes). Each putative homologous gene-pair is drawn as a gray dot on the dotplot with its x and y position corresponding to the genomic position of each gene in their respective genomes. Gene-pairs that have been inferred as syntenic (collinear order) are colored green. The collection of these dots appear as green line, which for the comparison of these two genomes, results in a nearly continuous green line running 45-degress up the dotplot. This means that these genomes are completely syntenic. |
| + | |||
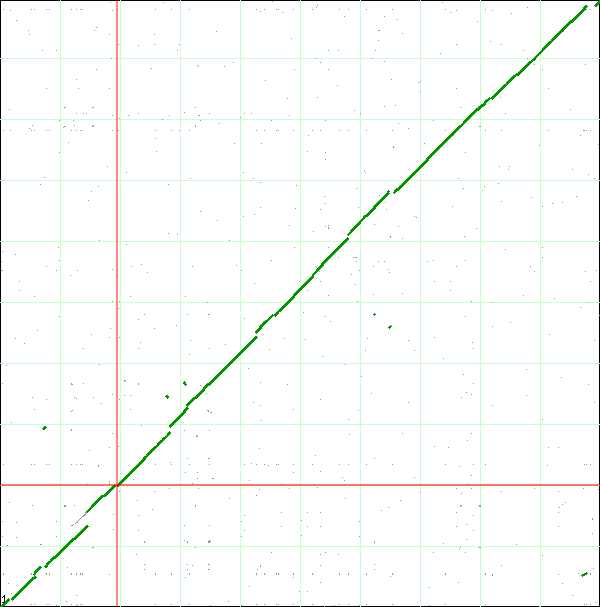
| + | [[Image:Dotplot.png|thumb|center|700px| Syntenic dotplot of Escherichia coli strain B REL606 and strain DH10B. The genomes are laid on the axes REL606 (x-axis) and DH10B (y-axis). The numbers correspond to the individual analysis of the "breaks" in the dotplot which could be found [[Analysis_of_variations_found_in_genomes_of_Escherichia_coli_strain_K12_DH10B_and_strain_B_REL606_using_SynMap_and_GEvo_analysis#Detailed_analysis_of_each_syntenic_discontinuity | here.]] ]] | ||
| + | |||
| + | If you look closely at the syntenic dotplot of these genomes, you'll notice that the syntenic line is not perfect, and there are many "breaks" or discontinuities between them. In the figure above, several these are indicated by a numbered arrow. These breaks in the syntenic path between these genomes are due to genomic changes happening at a larger scale than a single nucleotide polymorphism, and are mostly likely due to the insertion or deletion of a many nucleotide chunk in one of these genomes. In order to accurately account and characterize these discontinuities in the dotplot, you need to perform a high-resolution analysis of these regions using [[GEvo]]. | ||
| + | |||
| + | ==High-resolution sequence analysis using [[SynMap]]'s links to [[GEvo]]== | ||
| + | |||
| + | [[Image:Canvas1.png|thumb|center|700px| Red cross positioned at sixth break. Clicking here will open a new page of GEvo containing sequence information of both strains at this locus. Click "Run GEvo Analysis!" to visualize syntenic genes at this location]] | ||
| + | |||
| + | [[GEvo]] allows you to run pairwise comparisons between multiple genomic regions where you can specify how big of a genomic region to analyze. For more information on how to use [[GEvo]] please see its [[GEvo | help page.]] To analyse each of these "breaks" in the sytnenic dotplot, first click on the dotplot. This will open a new window with a close-up of the dotplot. While this is mostly a redundant function when comparing bacterial genomes, this features is important when dealing with genomes with multiple chromosomes. When this window appears use the cross-hair locator that appears when you mouse over the dotplot and place it on the green spot right before a "break". The locator will turn "red" when it is over a gene-pair that can be used as a link to GEvo. When the locator has turned red, click. For example, in order to visualize GEvo analysis of "break" number six, position the locator where the number six arrow points on dotplot. Click when the locator turns "red" or use this address: [http://tinyurl.com/ybokuag] to regenerate this particular analysis. | ||
| + | |||
| + | ==Running [[GEvo]]== | ||
| + | |||
| + | [[Image:GEvo.png|thumb|center|700px|Webpage of GEVo]] | ||
After positioning the locator and clicking , a new page for GEvo will appear displaying the sequence information corresponding to our region of interest in the dotplot. These sequences are "anchor" points into these two genomes for specifying the genomic regions to be compared. When linking to GEvo, SynMap automatically sets GEvo to specify using 50,000 nucleotides to the left and right of the anchor point. By default, GEvo will use BlastZ for its sequence comparison algorithm, which is a good choice for identifying large blocks of similar sequence. These settings (~100kb of each genome; BlastZ) usually work well for an initial analysis, and all you need to do is click "Run GEvo Analysis!". | After positioning the locator and clicking , a new page for GEvo will appear displaying the sequence information corresponding to our region of interest in the dotplot. These sequences are "anchor" points into these two genomes for specifying the genomic regions to be compared. When linking to GEvo, SynMap automatically sets GEvo to specify using 50,000 nucleotides to the left and right of the anchor point. By default, GEvo will use BlastZ for its sequence comparison algorithm, which is a good choice for identifying large blocks of similar sequence. These settings (~100kb of each genome; BlastZ) usually work well for an initial analysis, and all you need to do is click "Run GEvo Analysis!". | ||
| − | |||
| − | |||
| − | + | ==GEvo's results== | |
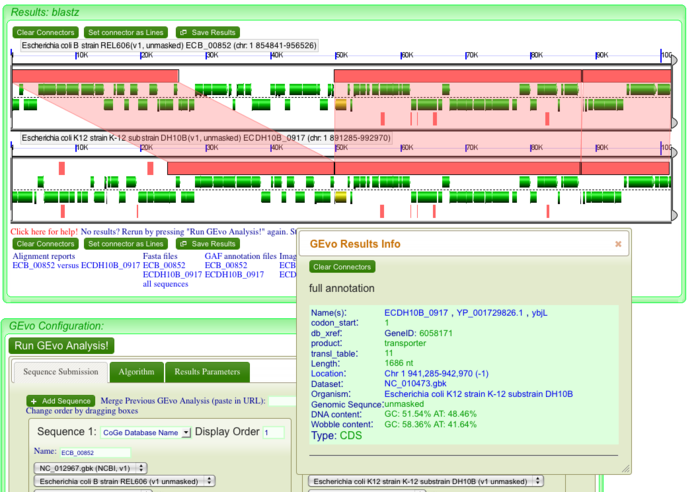
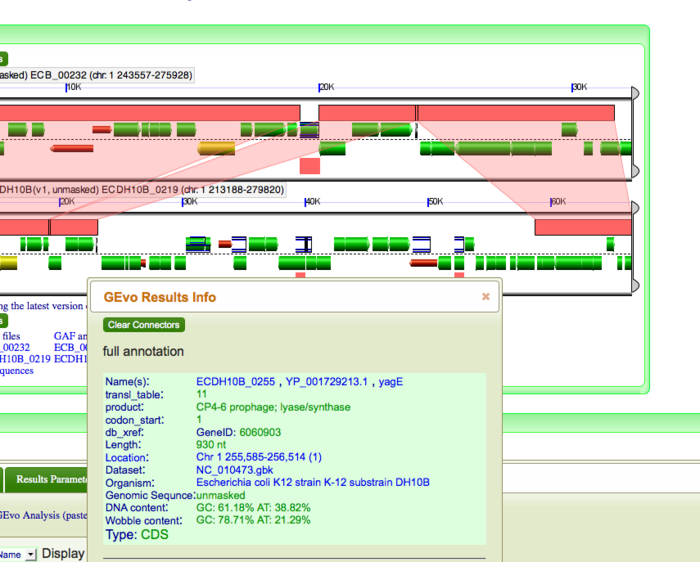
| + | Once the GEvo analysis results appear, we can begin to look for and characterize the differences between these two genomes at this syntenic region. GEvo's results will show two panels, one for each genomic region. The dashed line in the middle of each panel separates the top and bottom strands of DNA. Gene models are drawn as green arrows above and below this line, and clicking on a gene will cause its annotation to appear in a box. | ||
| − | + | [[Image:Gene annotation1.png|thumb|center|700px| To determine identity of genes, click on individual genes and its annotation will appear in a box]] | |
| − | + | The pink blocks in these panels are genomic regions identified by BlastZ as being similar in sequence composition. If you click on a pink block, a transparent wedge is drawn connecting it to its partner region in the other genome, and information about the blast hit (also known as an HSP) is shown in an information box. You can click on all the pink blocks individually to connect every region of sequence similarity, or hold the "shift" key and then click on one of the pink blocks to connect them all at once. Since we are analyzing "breaks" in our dotplot that are the result of an insertion or deletion, we expect to see at least one genomic region that is present in one genome and not the other. These indels will be evident by the pattern of transparent wedges and pink blocks. | |
| − | + | ==Zooming in on a region in [[GEvo]] | |
| + | [[Image:GEvo zooming in on region.png|center|thumb|700px| Using side bars zoom in on a region.]] | ||
| − | Let us consider the ninth break in the dotplot. Run [http://tinyurl.com/yk9kt7e GEvo Analysis] for this break. | + | As mentioned above, when you click on the dotplot, GEvo will display 50000 nucleotides towards the left and right from the anchor position. However, this may be display too much sequence. To zoom in on a region, use side bars to restrict the region of genome being displayed. This will magnify the region of interest and allows to view minute details. To get to this analysis directly, use this link: http://tinyurl.com/yemhyzg. |
| + | |||
| + | |||
| + | ==High-resolution [[GEvo]] analysis of an insertion: finding direct sequence repeats== | ||
| + | [[Image:GEvo-ecoli-insertion-direct-repeats.png|center|thumb|700px| Visualizing direct repeats around a putative insertion in the genome of Escherichia coli. Results can be regenerated at: http://tinyurl.com/yemhyzg ]] | ||
| + | |||
| + | Insertions in bacterial genome could be from exogenous DNA such as plasmids and phages or it could be the result of transposition or translocation. One way to gain evidence for a recent insertion is to look for [[direct repeats | direct repeated sequences]] boarding a putative inserions. These direct repeats are created at the site of insertion. Notice that on the edges of "inserted" genes in REL606, the pink wedges overlap at a common syntenic region in DH10B. This is visualized by the ends of the pink blocks overlapping slightly. These are direct repeats and evidence that an insertion happened in the genome of REL606. | ||
| + | |||
| + | To determine what the genes are in the inserion,just click on the gene models and their annotations will appear in a [[dialog box]] in [[GEvo]]. While many of the genes are annotated "hypothetical", several are annotated as phage genes. This is likely a prophage. | ||
| + | |||
| + | ==Differences at an insertion site== | ||
| + | Let us consider the ninth break in the dotplot. Run [http://tinyurl.com/yk9kt7e GEvo Analysis] for this break. | ||
| + | |||
| + | [[Image:Ecoli insertion with flagellar genes.png|thumb|center|700px|Putative insertion in DH10B containing many flagellar genes.]] | ||
| + | |||
| + | This is a different type of insertion event. In REL606 there is a single gene insertion, while in DH10B there is a many gene insertion at the same genomic position. In REL606, the gene is boarded by black-blue boxes, which is how annotated [[GenomeView_examples#Repeat_Regions | repeated sequences are visualized in CoGe]]. This gene is an IS1 [[transposon]], a class of DNA elements that move around a genome. The DH10B insertion contains a number of flagellar. These regions present some different possible evolutionary scenarios: | ||
| + | #Deletion in REL606: Perhaps two IS1 transposons landed in REL606 are were oriented in the same direction. This could provide [[direct repeat sequences]] needed for non-homologous recombination to remove the intervening sequence which included the flagellar genes | ||
| + | #Insertion in HD10B: Perhaps the flagellar genes were transferred in and replaced an IS1 element by hijacking its transposition machinery | ||
| + | #Insertion in both: Perhaps they are both new insertional events. | ||
| + | |||
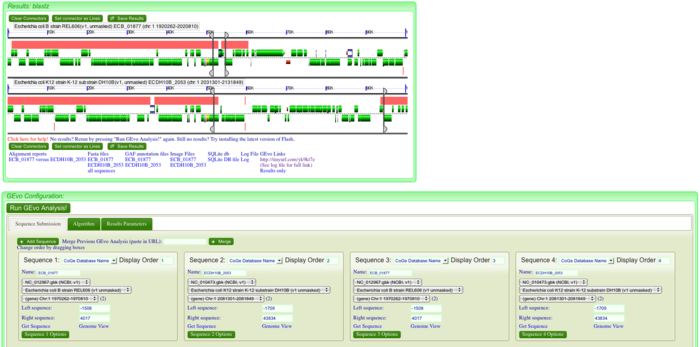
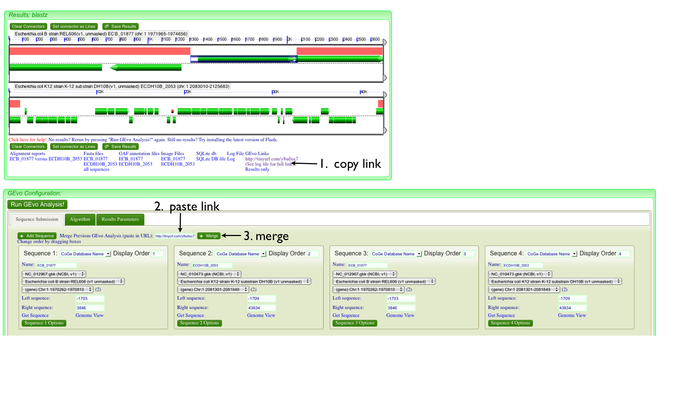
| + | Since the sequence at this position is not overlapping between the regions, we can investigate this by adding a second copy of each region to the analysis, and looking for repeat sequences boarding these putative insertions. There are two ways to do this: | ||
| + | |||
| + | [[Image: GEvo-4way-coli-setup.png|thumb|center|700px|Adding and resizing]] | ||
| + | #adding new sequences: | ||
| + | ##Click on "Add sequence" in GEvo | ||
| + | ##Copy and paste the gene name into the "name" box. Do this for each region. | ||
| + | ##Zoom in on the displayed sequences to use sequence right around the insertion and copy these positions in to the newly added sequences. | ||
| + | [[Image: GEvo-merge.002.png|thumb|center|700px|Adding and resizing]] | ||
| + | #Merge two analysis: | ||
| + | ##Zoom in on region | ||
| + | ##Run analysis | ||
| + | ##Copy link from analysis into merge box and press merge | ||
| + | |||
| + | The order of the sequence can be changed by dragging the sequence submission boxes around relative to one another. Also, the alignment algorithm should be changed to blastn instead of blastz. Blastn is more sensitive than blastz for finding small regions of sequence similarity. You can use this link to generate the results of this 4-way analysis: http://tinyurl.com/y9pgzvl . | ||
| + | |||
| + | [[Image:GEvo-4way-coli-insertion.png|thumb|center|700px|4-way GEvo analysis including self-self comparisons. http://tinyurl.com/y9pgzvl]] | ||
| + | |||
| + | The results from this 4-way analysis do not show any direct sequence repeats at the ends of either insertion. This means that there is not evidence that region in DH10B was inserted recently. Of the possible evolutionary scenarios, it is most likely that this region was deleted in REL606, probably as the result of the IS1 element, perhaps by the insertion of two of these elements followed by deletion of the intervening sequence. | ||
| + | |||
| + | ==Phage insertion== | ||
| + | |||
| + | |||
| + | |||
| + | Next, we will look at phage insertion. Consider the second break in the dotplot. Run [http://tinyurl.com/yjdqgzr GEVo analysis] on this break. Before checking for direct repeats, determine the identity of genes inserted in DH10B. These are phage-specific genes. CP4-6 prophage has integrated its DNA at this locus as seen by CP4-6 specific integrase, DNA binding protein etc. [[Image:phage.png|thumb|center|700px |In DH10B, the inserted genes are CP4-6 prophage specific as seen by gene annotations.]] | ||
| + | |||
| + | Next we will look at an example of inversion. Consider the twelfth break on the dotplot. Run [http://tinyurl.com/yhyxgrq GEvo Analysis] on this break. Notice the syntenic regions between 10K and 30K on DH10B. Click on the pink blocks and notice the pattern of transparent wedges that connect the syntenic regions. These genes are inverted. Notice IS10R and IS10L bordering the inverted region. These IS elements have created inverted terminal repeats (ITR) which tend to invert or flip the genes within them. | ||
| + | [[Image:Inversion.png|thumb|center|700px| In DH10B, the insertion of IS10R and IS10L has created inverted terminal repeats i.e the IS10 (IS10R and IS10L) transposons are integrated in opposite orientations. A cross over between these two transposons has inverted the DNA segment(three genes) within it. Notice the patterns of wedges connecting the syntenic region (they cross each other). Use side bars to better visualize this inversion event]] | ||
| + | |||
| + | ==Detailed analysis of each syntenic discontinuity== | ||
| + | [[Image:Dotplot.png|thumb|right|600px| Syntenic dotplot of Escherichia coli strain B REL606 and strain DH10B. The genomes are laid on the axes REL606 (x-axis) and DH10B (y-axis). The numbers correspond to the individual analysis of the "breaks" in the dotplot.]] | ||
| + | {| width="1000" cellspacing="1" cellpadding="1" border="1" | ||
| + | |- | ||
| + | | Variation type<br> | ||
| + | | Difference in strain B REL606<br> | ||
| + | | Difference in strain K-12 DH10B<br> | ||
| + | | Evidence<br> | ||
| + | | Notes<br> | ||
| + | | Link leading to GEvo <br> | ||
| + | |- | ||
| + | | 1. Deletion<br> | ||
| + | | none<br> | ||
| + | | Deletion of ~18 genes including DNA <br>pol II, genes in metabolic pathway, thiamine ABC transporter<br><br> | ||
| + | | pseudogenes in DH10B at deletion site.<br><br> | ||
| + | | Possible additional insertion in DH10B as evidenced by <br>pseudogenes of yabP, RNA pol associated helicase and FruR, that are not present in REl606<br><br> | ||
| + | | [http://tinyurl.com/ylg9qrk tinyurl.com/yexrzpb]<br> | ||
| + | |- | ||
| + | | 2. Insertion<br> | ||
| + | | Insertion of IS1 transposon<br> | ||
| + | | Insertion sequences and Prophage CP46 DNA insertion | ||
| + | | Prophage specific genes found in DH10B<br> | ||
| + | | Prophage DNA insertion and IS insertions has created pseudogenes in K-12 DH10B<br> | ||
| + | | [http://tinyurl.com/yjdqgzr tinyurl.com/yd2quy7]<br> | ||
| + | |- | ||
| + | | 3. Translocation in REL606 and insertion in DH10B<br> | ||
| + | | Insertion of IS1 sequence. Translocation of ~15 genes including lac operon and other metabolic enzymes genes | ||
| + | | Insertion of IS3 and IS2 sequences | ||
| + | | | ||
| + | Translocation in REL606 as evidenced by direct repeats.Dotplot shows that the missing genes are present in DH10B but not in this locus. The syntenic region is therefore not colinear.<br> | ||
| + | |||
| + | | | ||
| + | Pseudogenes of yaiT and yaiX were created in DH10B by transposon insertions. | ||
| + | |||
| + | Insertion by translocation in REL606 was confirmed as lac operon and other metabolic genes were found in DH10B by analyzing the translocated genes on the dotplot | ||
| + | |||
| + | | [http://tinyurl.com/yldc83u http://tinyurl.com/yldc83u]<br> | ||
| + | |- | ||
| + | | 4. Insertion in REL606 and DNA duplication event in DH10B. <br> | ||
| + | | Prophage DNA and transposase insertion <br> | ||
| + | | Recent DNA duplication event | ||
| + | | 100% identity between paralogs in DH10B and ~98% identity between syntenic region of DH10B and REL606<br> | ||
| + | | Possible phage DNA insertion in REL606 as "hypothetical protein" genes were found near putative prophage tail component gene in REL606. <br> | ||
| + | | [http://tinyurl.com/yk7vjgq tinyurl.com/yea8bu6]<br> | ||
| + | |- | ||
| + | | 5. Insertion<br> | ||
| + | | Bacteriophage DNA insertion <br> | ||
| + | | IS2 sequence insertion<br> | ||
| + | | Pseudogenes at IS2 insertion site in DH10B. Phage specific genes were found in REL606<br> | ||
| + | | Possible phage DNA insertion in REL606 as "Hypothetical proteins" were found near phage specific genes <br> | ||
| + | | [http://tinyurl.com/yevlb2w tinyurl.com/yevlb2w]<br> | ||
| + | |- | ||
| + | | 6. Insertion<br> | ||
| + | | Prophage DNA insertion <br> | ||
| + | | none<br> | ||
| + | | Phage specific genes were found in REL606<br> | ||
| + | | none<br> | ||
| + | | [http://tinyurl.com/ybokuag tinyurl.com/ybokuag]<br> | ||
| + | |- | ||
| + | | 7. Insertion,translocation and inversion<span class="Apple-tab-span" style="white-space: pre;"> </span><br> | ||
| + | | none | ||
| + | | Prophage DNA insertion and translocation of nitrite reductase 2 genes | ||
| + | | Phage specific genes found in DH10B<br> | ||
| + | | Translocation in DH10B is evident by dotplot. Moreover, the translocated genes in DH10B were found to be inverted. It could not be determined genes on which genomes were inverted as tranposon insertions were found in both genomes.<br> | ||
| + | | | ||
| + | [http://tinyurl.com/yaxlh7o tinyurl.com/yaxlh7o] | ||
| + | |||
| + | [http://tinyurl.com/y9cs6ft http://tinyurl.com/y9cs6ft] | ||
| + | |||
| + | |- | ||
| + | | 8. Insertion and deletion<br> | ||
| + | | Transposon insertions and deletion of phenylacetic acid degradation genes <br> | ||
| + | | IS and Rac prophage DNA insertion | ||
| + | | Phage specific genes found in DH10B. IS or transposon insertions in REL606 might have created direct repeats and facilitated excision of phenylacetic acid degradation genes.<br> | ||
| + | | Rac prophage DNA disrupted by transposon insertion in DH10B | ||
| + | | [http://tinyurl.com/ylgv7xc tinyurl.com/yccbmsq]<br> | ||
| + | |- | ||
| + | | 9a. Insertion | ||
| + | | 9a. none<span class="Apple-tab-span" style="white-space: pre;"> </span> | ||
| + | | 9a. Insertion of IS5 sequence | ||
| + | | 9a. none | ||
| + | | 9a. none | ||
| + | | 9a. [http://tinyurl.com/ylllc6u http://tinyurl.com/ylllc6u] | ||
| + | |- | ||
| + | | 9b. Insertion | ||
| + | | 9b. Insertion of ISI transposon | ||
| + | | 9b. none | ||
| + | | 9b. none | ||
| + | | 9b. none | ||
| + | | 9b. [http://tinyurl.com/ygsqg2f tinyurl.com/ygsqg2f] | ||
| + | |- | ||
| + | | 9c. Insertion | ||
| + | | 9c. ISI insertion<br> | ||
| + | | 9c. Insertion of ABC transporter, flagella encoding genes and few other enzymes | ||
| + | | 9c. Inserted DNA segment in DH10B is bordered by direct repeats at both ends. 100% identity was found between the two repeats. <br> | ||
| + | | 9c.DR indicates transposon insertion in DH10B. | ||
| + | | 9c[http://tinyurl.com/yza4jy3 tinyurl.com/yza4jy3][http://tinyurl.com/yjlns3p] | ||
| + | |- | ||
| + | | 9d. Insertion | ||
| + | | 9d. IS2 insertion <br> | ||
| + | | 9d. none | ||
| + | | 9d. none | ||
| + | | 9d. none | ||
| + | | 9d. [http://tinyurl.com/ygfgtqy tinyurl.com/ygfgtqy] | ||
| + | |- | ||
| + | | 10. Insertion and deletion<br><br> | ||
| + | | Bacteriophage DNA insertion and IS1 transposon insertion. Deletion of ~5 genes<br><br> | ||
| + | | Insertion of IS3<br> | ||
| + | | IS1 insertion at the site of deletion. Another IS1 insertion might have created direct repeats and facilitated deletion. | ||
| + | | none | ||
| + | | [http://tinyurl.com/ykynub2 tinyurl.com/ykynub2]<br> | ||
| + | |- | ||
| + | | 11. Insertion and deletion <br> | ||
| + | | IS1 insertion<br> | ||
| + | | CP4-57 prophage DNA insertion and possible deletion of ParB family protein and recombinase<br> | ||
| + | | Phage insertion in DH10B may have created pseudogene of ParB family protein genes and recombinase which later got deleted <br> | ||
| + | | Pseudogenes of yqa, yga and ypj indicated possible formation of pseudogenes of ParB and recombinase at some time prior to their deletion in DH10B.<br> | ||
| + | | [http://tinyurl.com/yg7ybg4 tinyurl.com/yg7ybg4]<br> | ||
| + | |- | ||
| + | | 12. Insertion, deletion and Inversion<br> | ||
| + | | IS1 insertion. | ||
| + | | IS5 and IS10 transposon insertion. Inversion of ornithine decarboxylase, M-type protein and bifunctional prepilin peptidase/methylase. Deletion of saframycin synthetase, capsule related genes, bio-film formation genes, anti-toxin system and type II secretory apparatus genes. | ||
| + | | | ||
| + | Inversion in DH10B as evidenced by inverted repeats of IS10 transposon. | ||
| + | |||
| + | Deletion of several genes in DH10B is evidenced by IS5 trans-activator transposase and presence of pseudogenes in DH10B, | ||
| + | |||
| + | | | ||
| + | Insertion of IS5 trans-activator transposase indicates possible deletion of several genes in DH10B. Also, no evidence of insertion in REL606 was found such as direct repeats. | ||
| + | |||
| + | | [http://tinyurl.com/yhyxgrq tinyurl.com/yhyxgrq]<br> | ||
| + | |- | ||
| + | | | ||
| + | 13a. Deletion | ||
| + | |||
| + | | 13a. none | ||
| + | | | ||
| + | 13a. Deletion of putative adhesin<br> | ||
| + | |||
| + | | 13a. No direct repeats were found to indicate insertion of putative adhensin in REL606 therefore deletion in DH10B may have happened | ||
| + | | 13a.none<br> | ||
| + | | 13a.[http://tinyurl.com/yjojy53 tinyurl.com/yjojy53] | ||
| + | |- | ||
| + | | | ||
| + | 13b. Insertion<br> | ||
| + | |||
| + | | 13b. IS1 insertion and deletion of lipopolysaccharide genes | ||
| + | | | ||
| + | 13b. none<br> | ||
| + | |||
| + | | | ||
| + | <br> 13b. IS1 insertion in REL606 indicates that deletion may have occured by formation of directed repeats. | ||
| + | |||
| + | | 13b. IS1 insertion created pseudogene. | ||
| + | | 13b.[http://tinyurl.com/yj2yg5s http://tinyurl.com/yj2yg5s] | ||
| + | |- | ||
| + | | | ||
| + | 13c. Insertion | ||
| + | |||
| + | and deletion<br> | ||
| + | |||
| + | | | ||
| + | 13c. Insertion of IS30 transposon and several 'hypothetical protein" genes. | ||
| + | |||
| + | | | ||
| + | 13c. none<br> | ||
| + | |||
| + | | | ||
| + | 13c. Insertion in REL606 is evidenced by direct repeats<br> | ||
<br> | <br> | ||
| + | | | ||
<br> | <br> | ||
| − | + | 13c. direct repeats were found in REL606 which indicates insertion of ShiA-like and TrbC-like genes. <br> | |
| − | + | | | |
| − | + | <br> | |
| − | + | ||
<br> | <br> | ||
| − | + | 13c.[http://tinyurl.com/yjzdyum tinyurl.com/yjzdyum]<br> | |
| − | + | [http://tinyurl.com/ydkrcv8 <br>] | |
| − | + | |- | |
| + | | | ||
| + | 14a. Insertion | ||
| − | + | | 14a. Insertion of several transposons and secondary glycine betaine transporter | |
| + | | 14a.Insertion of several transposons. Insertion of Kple2 phage-like element | ||
| + | | 14a. Direct repeats bordering secondary glycine betaine transporter indicates its insertion | ||
| + | | 14a. none | ||
| + | | 14a. [http://tinyurl.com/yzyvunx tinyurl.com/yzyvunx] | ||
| + | |- | ||
| + | | 14b. Insertion | ||
| + | | 14b. Insertion of ~15 genes | ||
| + | | 14b. Phage insertion. Transposon insertions | ||
| + | | 14b. Insertion in REL606 is evidenced by direct repeats flanking the DNA segment containing several genes. | ||
| + | | 14b.Phage-like genes were found in DH10B | ||
| + | | 14b. [http://tinyurl.com/yly2b6u tinyurl.com/yly2b6u] | ||
| + | |- | ||
| + | | | ||
| + | <br> 14c. Deletion | ||
| − | + | | | |
| + | 14c. none | ||
| − | + | | | |
| + | <br> | ||
| − | + | 14c. Deletion of ~15 genes. | |
| − | + | <br> | |
| − | + | <br> | |
| + | | | ||
<br> | <br> | ||
| − | + | 14c. Deletion in DH10B is evidenced by insertion of IS10R which may have facilitated excision of DNA by forming direct repeats | |
| − | + | | | |
| + | <br> | ||
| − | <br> | + | <br> |
| − | + | 14c. Pseudogenes found at the site of deletion and IS10R insertion. | |
| + | |||
| + | | | ||
| + | <br> | ||
| + | |||
| + | <br> | ||
| − | [ | + | 14c. [http://tinyurl.com/yfhhsk6 tinyurl.com/yfhhsk6] |
| − | <br> | + | [http://tinyurl.com/ycwsmsl <br>] |
| − | + | |} | |
| − | + | <br> | |
Latest revision as of 15:19, 12 February 2010
Contents
- 1 Background
- 2 Generating a syntenic dotplot of two Escherichia coli strains
- 3 Syntenic dotplot data interpretation and analysis
- 4 High-resolution sequence analysis using SynMap's links to GEvo
- 5 Running GEvo
- 6 GEvo's results
- 7 High-resolution GEvo analysis of an insertion: finding direct sequence repeats
- 8 Differences at an insertion site
- 9 Phage insertion
- 10 Detailed analysis of each syntenic discontinuity
Background
In this exercise you will compare the genomes of two Escherichia coli strains, K12 DH10B and B REL606, using whole genome syntenic comparison and high-resolution analyses of specific genomic regions. These analyses will use CoGe's tools SynMap and GEvo respectively, and will reveal evolutionary changes between these two genomes that happened after the divergence of their lineages. While the nucleotide sequence of these genomes is identical over large expanses of their genomes, many other types of large-scale genomic change will be discovered including phage insertions, transposon transposition, and genomic insertion, deletion, inversion, and duplication events. The computational tools used to do these analyses can be used for comparing the genomes of any organisms. To learn what organisms and genomes are available in CoGe, please see GenomeView.
Generating a syntenic dotplot of two Escherichia coli strains
Synteny, in genomic terms, is defined as two or more genomic regions that are derived from a common ancestor. To identify syntenic regions, you are going generate a syntenic dotplot of the genomes of Echerichia coli strains K12 DH10B and B REL606 using SynMap. First, go to SynMap. Search for these E. coli strains in CoGe's database by typing in part of the their names in the "Name" search boxes for Organism 1 and Organism 2. For example, search for "DH10B" and "REL606" respectively, or type "escheri" in both boxes. Alternatively, use this link to load SynMap with these genomes already specified. Once SynMap has found organisms that matches these names, make sure they are selected in the Organism List. While there are several parameters that can be configured when generating a syntenic dotplot using SynMap, the default settings work well for most situations, and very well for closely related organisms. Click "Generate SynMap" to start the analysis.
A lot of processing is happening behind the scenes, but the general way a syntenic dotplot is created is:
- All protein coding regions (CDS)) are extracted from each genome
- These sequences are blasted against each other to identify putative homologous gene pairs
- Putative homologous gene pairs are analyzed to determine if they share a collinear order between the genoems
The general principle is that the most likely and parsimonious way two genomes have a collinear series of homologous genes is those genomic regions in each organism are derived from a common ancestral genomic region (hence they are syntenic). In other words, genomic synteny is inferred by a collinear arrangement of putatively homologous genes in two or more genomic genomic regions.
Syntenic dotplot data interpretation and analysis
When finished, SynMap will display a dotplot. Each axis of the dotplot is in nucleotide units and represents one of the two genomes laid end to end. The lower-left corner represents the start of each genome (usually 'ORI' for circular bacterial genomes). Each putative homologous gene-pair is drawn as a gray dot on the dotplot with its x and y position corresponding to the genomic position of each gene in their respective genomes. Gene-pairs that have been inferred as syntenic (collinear order) are colored green. The collection of these dots appear as green line, which for the comparison of these two genomes, results in a nearly continuous green line running 45-degress up the dotplot. This means that these genomes are completely syntenic.
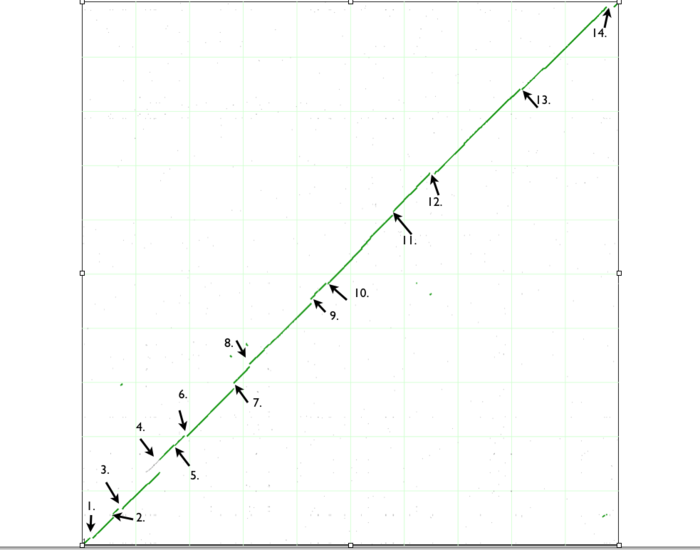
If you look closely at the syntenic dotplot of these genomes, you'll notice that the syntenic line is not perfect, and there are many "breaks" or discontinuities between them. In the figure above, several these are indicated by a numbered arrow. These breaks in the syntenic path between these genomes are due to genomic changes happening at a larger scale than a single nucleotide polymorphism, and are mostly likely due to the insertion or deletion of a many nucleotide chunk in one of these genomes. In order to accurately account and characterize these discontinuities in the dotplot, you need to perform a high-resolution analysis of these regions using GEvo.
High-resolution sequence analysis using SynMap's links to GEvo
GEvo allows you to run pairwise comparisons between multiple genomic regions where you can specify how big of a genomic region to analyze. For more information on how to use GEvo please see its help page. To analyse each of these "breaks" in the sytnenic dotplot, first click on the dotplot. This will open a new window with a close-up of the dotplot. While this is mostly a redundant function when comparing bacterial genomes, this features is important when dealing with genomes with multiple chromosomes. When this window appears use the cross-hair locator that appears when you mouse over the dotplot and place it on the green spot right before a "break". The locator will turn "red" when it is over a gene-pair that can be used as a link to GEvo. When the locator has turned red, click. For example, in order to visualize GEvo analysis of "break" number six, position the locator where the number six arrow points on dotplot. Click when the locator turns "red" or use this address: [1] to regenerate this particular analysis.
Running GEvo
After positioning the locator and clicking , a new page for GEvo will appear displaying the sequence information corresponding to our region of interest in the dotplot. These sequences are "anchor" points into these two genomes for specifying the genomic regions to be compared. When linking to GEvo, SynMap automatically sets GEvo to specify using 50,000 nucleotides to the left and right of the anchor point. By default, GEvo will use BlastZ for its sequence comparison algorithm, which is a good choice for identifying large blocks of similar sequence. These settings (~100kb of each genome; BlastZ) usually work well for an initial analysis, and all you need to do is click "Run GEvo Analysis!".
GEvo's results
Once the GEvo analysis results appear, we can begin to look for and characterize the differences between these two genomes at this syntenic region. GEvo's results will show two panels, one for each genomic region. The dashed line in the middle of each panel separates the top and bottom strands of DNA. Gene models are drawn as green arrows above and below this line, and clicking on a gene will cause its annotation to appear in a box.
The pink blocks in these panels are genomic regions identified by BlastZ as being similar in sequence composition. If you click on a pink block, a transparent wedge is drawn connecting it to its partner region in the other genome, and information about the blast hit (also known as an HSP) is shown in an information box. You can click on all the pink blocks individually to connect every region of sequence similarity, or hold the "shift" key and then click on one of the pink blocks to connect them all at once. Since we are analyzing "breaks" in our dotplot that are the result of an insertion or deletion, we expect to see at least one genomic region that is present in one genome and not the other. These indels will be evident by the pattern of transparent wedges and pink blocks.
==Zooming in on a region in GEvo
As mentioned above, when you click on the dotplot, GEvo will display 50000 nucleotides towards the left and right from the anchor position. However, this may be display too much sequence. To zoom in on a region, use side bars to restrict the region of genome being displayed. This will magnify the region of interest and allows to view minute details. To get to this analysis directly, use this link: http://tinyurl.com/yemhyzg.
High-resolution GEvo analysis of an insertion: finding direct sequence repeats
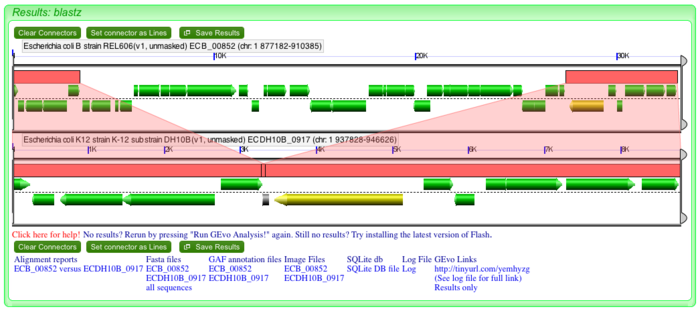
Insertions in bacterial genome could be from exogenous DNA such as plasmids and phages or it could be the result of transposition or translocation. One way to gain evidence for a recent insertion is to look for direct repeated sequences boarding a putative inserions. These direct repeats are created at the site of insertion. Notice that on the edges of "inserted" genes in REL606, the pink wedges overlap at a common syntenic region in DH10B. This is visualized by the ends of the pink blocks overlapping slightly. These are direct repeats and evidence that an insertion happened in the genome of REL606.
To determine what the genes are in the inserion,just click on the gene models and their annotations will appear in a dialog box in GEvo. While many of the genes are annotated "hypothetical", several are annotated as phage genes. This is likely a prophage.
Differences at an insertion site
Let us consider the ninth break in the dotplot. Run GEvo Analysis for this break.
This is a different type of insertion event. In REL606 there is a single gene insertion, while in DH10B there is a many gene insertion at the same genomic position. In REL606, the gene is boarded by black-blue boxes, which is how annotated repeated sequences are visualized in CoGe. This gene is an IS1 transposon, a class of DNA elements that move around a genome. The DH10B insertion contains a number of flagellar. These regions present some different possible evolutionary scenarios:
- Deletion in REL606: Perhaps two IS1 transposons landed in REL606 are were oriented in the same direction. This could provide direct repeat sequences needed for non-homologous recombination to remove the intervening sequence which included the flagellar genes
- Insertion in HD10B: Perhaps the flagellar genes were transferred in and replaced an IS1 element by hijacking its transposition machinery
- Insertion in both: Perhaps they are both new insertional events.
Since the sequence at this position is not overlapping between the regions, we can investigate this by adding a second copy of each region to the analysis, and looking for repeat sequences boarding these putative insertions. There are two ways to do this:
- adding new sequences:
- Click on "Add sequence" in GEvo
- Copy and paste the gene name into the "name" box. Do this for each region.
- Zoom in on the displayed sequences to use sequence right around the insertion and copy these positions in to the newly added sequences.
- Merge two analysis:
- Zoom in on region
- Run analysis
- Copy link from analysis into merge box and press merge
The order of the sequence can be changed by dragging the sequence submission boxes around relative to one another. Also, the alignment algorithm should be changed to blastn instead of blastz. Blastn is more sensitive than blastz for finding small regions of sequence similarity. You can use this link to generate the results of this 4-way analysis: http://tinyurl.com/y9pgzvl .
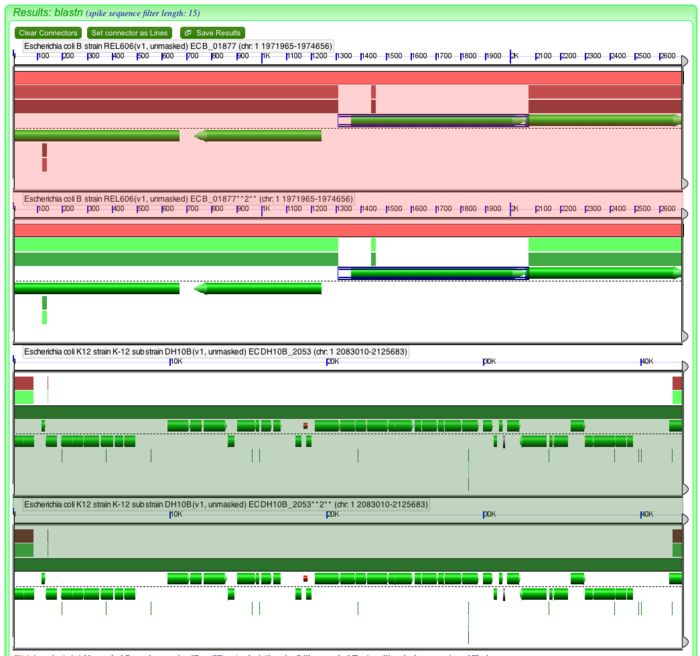
The results from this 4-way analysis do not show any direct sequence repeats at the ends of either insertion. This means that there is not evidence that region in DH10B was inserted recently. Of the possible evolutionary scenarios, it is most likely that this region was deleted in REL606, probably as the result of the IS1 element, perhaps by the insertion of two of these elements followed by deletion of the intervening sequence.
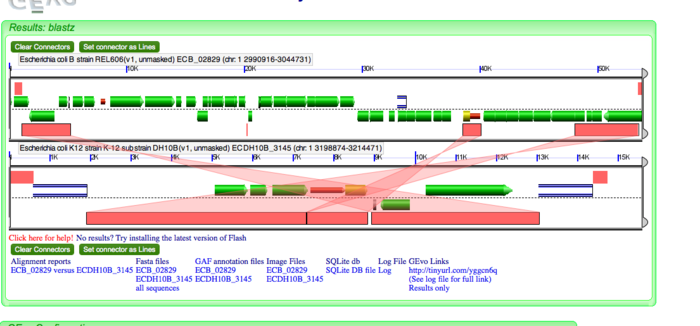
Phage insertion
Next, we will look at phage insertion. Consider the second break in the dotplot. Run GEVo analysis on this break. Before checking for direct repeats, determine the identity of genes inserted in DH10B. These are phage-specific genes. CP4-6 prophage has integrated its DNA at this locus as seen by CP4-6 specific integrase, DNA binding protein etc.Next we will look at an example of inversion. Consider the twelfth break on the dotplot. Run GEvo Analysis on this break. Notice the syntenic regions between 10K and 30K on DH10B. Click on the pink blocks and notice the pattern of transparent wedges that connect the syntenic regions. These genes are inverted. Notice IS10R and IS10L bordering the inverted region. These IS elements have created inverted terminal repeats (ITR) which tend to invert or flip the genes within them.
Detailed analysis of each syntenic discontinuity
| Variation type |
Difference in strain B REL606 |
Difference in strain K-12 DH10B |
Evidence |
Notes |
Link leading to GEvo |
| 1. Deletion |
none |
Deletion of ~18 genes including DNA pol II, genes in metabolic pathway, thiamine ABC transporter |
pseudogenes in DH10B at deletion site. |
Possible additional insertion in DH10B as evidenced by pseudogenes of yabP, RNA pol associated helicase and FruR, that are not present in REl606 |
tinyurl.com/yexrzpb |
| 2. Insertion |
Insertion of IS1 transposon |
Insertion sequences and Prophage CP46 DNA insertion | Prophage specific genes found in DH10B |
Prophage DNA insertion and IS insertions has created pseudogenes in K-12 DH10B |
tinyurl.com/yd2quy7 |
| 3. Translocation in REL606 and insertion in DH10B |
Insertion of IS1 sequence. Translocation of ~15 genes including lac operon and other metabolic enzymes genes | Insertion of IS3 and IS2 sequences |
Translocation in REL606 as evidenced by direct repeats.Dotplot shows that the missing genes are present in DH10B but not in this locus. The syntenic region is therefore not colinear. |
Pseudogenes of yaiT and yaiX were created in DH10B by transposon insertions. Insertion by translocation in REL606 was confirmed as lac operon and other metabolic genes were found in DH10B by analyzing the translocated genes on the dotplot |
http://tinyurl.com/yldc83u |
| 4. Insertion in REL606 and DNA duplication event in DH10B. |
Prophage DNA and transposase insertion |
Recent DNA duplication event | 100% identity between paralogs in DH10B and ~98% identity between syntenic region of DH10B and REL606 |
Possible phage DNA insertion in REL606 as "hypothetical protein" genes were found near putative prophage tail component gene in REL606. |
tinyurl.com/yea8bu6 |
| 5. Insertion |
Bacteriophage DNA insertion |
IS2 sequence insertion |
Pseudogenes at IS2 insertion site in DH10B. Phage specific genes were found in REL606 |
Possible phage DNA insertion in REL606 as "Hypothetical proteins" were found near phage specific genes |
tinyurl.com/yevlb2w |
| 6. Insertion |
Prophage DNA insertion |
none |
Phage specific genes were found in REL606 |
none |
tinyurl.com/ybokuag |
| 7. Insertion,translocation and inversion |
none | Prophage DNA insertion and translocation of nitrite reductase 2 genes | Phage specific genes found in DH10B |
Translocation in DH10B is evident by dotplot. Moreover, the translocated genes in DH10B were found to be inverted. It could not be determined genes on which genomes were inverted as tranposon insertions were found in both genomes. |
|
| 8. Insertion and deletion |
Transposon insertions and deletion of phenylacetic acid degradation genes |
IS and Rac prophage DNA insertion | Phage specific genes found in DH10B. IS or transposon insertions in REL606 might have created direct repeats and facilitated excision of phenylacetic acid degradation genes. |
Rac prophage DNA disrupted by transposon insertion in DH10B | tinyurl.com/yccbmsq |
| 9a. Insertion | 9a. none | 9a. Insertion of IS5 sequence | 9a. none | 9a. none | 9a. http://tinyurl.com/ylllc6u |
| 9b. Insertion | 9b. Insertion of ISI transposon | 9b. none | 9b. none | 9b. none | 9b. tinyurl.com/ygsqg2f |
| 9c. Insertion | 9c. ISI insertion |
9c. Insertion of ABC transporter, flagella encoding genes and few other enzymes | 9c. Inserted DNA segment in DH10B is bordered by direct repeats at both ends. 100% identity was found between the two repeats. |
9c.DR indicates transposon insertion in DH10B. | 9ctinyurl.com/yza4jy3[2] |
| 9d. Insertion | 9d. IS2 insertion |
9d. none | 9d. none | 9d. none | 9d. tinyurl.com/ygfgtqy |
| 10. Insertion and deletion |
Bacteriophage DNA insertion and IS1 transposon insertion. Deletion of ~5 genes |
Insertion of IS3 |
IS1 insertion at the site of deletion. Another IS1 insertion might have created direct repeats and facilitated deletion. | none | tinyurl.com/ykynub2 |
| 11. Insertion and deletion |
IS1 insertion |
CP4-57 prophage DNA insertion and possible deletion of ParB family protein and recombinase |
Phage insertion in DH10B may have created pseudogene of ParB family protein genes and recombinase which later got deleted |
Pseudogenes of yqa, yga and ypj indicated possible formation of pseudogenes of ParB and recombinase at some time prior to their deletion in DH10B. |
tinyurl.com/yg7ybg4 |
| 12. Insertion, deletion and Inversion |
IS1 insertion. | IS5 and IS10 transposon insertion. Inversion of ornithine decarboxylase, M-type protein and bifunctional prepilin peptidase/methylase. Deletion of saframycin synthetase, capsule related genes, bio-film formation genes, anti-toxin system and type II secretory apparatus genes. |
Inversion in DH10B as evidenced by inverted repeats of IS10 transposon. Deletion of several genes in DH10B is evidenced by IS5 trans-activator transposase and presence of pseudogenes in DH10B, |
Insertion of IS5 trans-activator transposase indicates possible deletion of several genes in DH10B. Also, no evidence of insertion in REL606 was found such as direct repeats. |
tinyurl.com/yhyxgrq |
|
13a. Deletion |
13a. none |
13a. Deletion of putative adhesin |
13a. No direct repeats were found to indicate insertion of putative adhensin in REL606 therefore deletion in DH10B may have happened | 13a.none |
13a. tinyurl.com/yjojy53 |
|
13b. Insertion |
13b. IS1 insertion and deletion of lipopolysaccharide genes |
13b. none |
|
13b. IS1 insertion created pseudogene. | 13b.http://tinyurl.com/yj2yg5s |
|
13c. Insertion and deletion |
13c. Insertion of IS30 transposon and several 'hypothetical protein" genes. |
13c. none |
13c. Insertion in REL606 is evidenced by direct repeats
|
13c. direct repeats were found in REL606 which indicates insertion of ShiA-like and TrbC-like genes. |
13c. tinyurl.com/yjzdyum |
|
14a. Insertion |
14a. Insertion of several transposons and secondary glycine betaine transporter | 14a.Insertion of several transposons. Insertion of Kple2 phage-like element | 14a. Direct repeats bordering secondary glycine betaine transporter indicates its insertion | 14a. none | 14a. tinyurl.com/yzyvunx |
| 14b. Insertion | 14b. Insertion of ~15 genes | 14b. Phage insertion. Transposon insertions | 14b. Insertion in REL606 is evidenced by direct repeats flanking the DNA segment containing several genes. | 14b.Phage-like genes were found in DH10B | 14b. tinyurl.com/yly2b6u |
|
|
14c. none |
14c. Deletion of ~15 genes.
|
14c. Deletion in DH10B is evidenced by insertion of IS10R which may have facilitated excision of DNA by forming direct repeats |
14c. Pseudogenes found at the site of deletion and IS10R insertion. |
14c. tinyurl.com/yfhhsk6 |