How to extract genomic features
Contents
- 1 Overview
- 2 Finding the genomic region of interest
- 3 Visualizing a genomic region of interest in GenomeView and exporting the genomic features from a genomic region
- 4 Evaluating, selecting, and exporting genomic features in FeatList
- 5 Displaying Fasta formatted sequences in FastaView
- 6 Congratulations! You have learned how to extract genomic features from a genomic region!
Overview
Extracting genomic features from a genomic region in a genome of interest is easy using GenomeView. All you need to do is:
- Find your genome of interest in OrganismView or your genomic feature in FeatView
- Visualize the genomic region of interest in GenomeView
- Select and export a genomic region from OrganismView for automatic feature extraction using FeatList
- Select the features of interest and send them to FastaView to get their sequences in fasta format
Finding the genomic region of interest
There are two general ways to find a genomic region of interest depending on if you know:
- The genomic location by organism, chromosome, and nucleotide position
- The name of a genomic feature in the genomic region
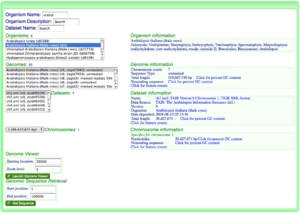
FeatView's results after searching for genomic features with the name "At1g01010". The Arabidopsis thaliana genome from TAIR version 9 is selected and the feature type "CDS" is selected.
Find your genome of interest in OrganismView
This option should be used if you know the organism, chromosome, and nucleotide position of the genomic region in which you are interested.
- Go to OrganismView and search for your organism by name by typing part of its name in the name search box, or part of the organism description in the description box. For more information about this, please see OrganismView.
- Select the correct organism from the organism list.
- Select the appropriate genome from the list of genomes for that organism.
- If needed, select the appropriate dataset and chromosome for the genome.
- Type in the appropriate location of the genomic region in which you are interested in the "Starting location" box under the "Genome Viewer" section in the lower right of the screen of OrganismView.
- Press the button called "Launch Genome Viewer" to launch GenomeView to display this genomic region.
Find your genomic feature in FeatView
This option should be used if you know the name of the genomic feature in which you are interested.
- Go to FeatView and search for your genomic feature by name or annotation. For more information about this, please see FeatView.
- Select the correct genome, dataset and genomic feature type for your genomic feature.
- Press the "Genome Browser" button located above the displayed annotations for the selected genomic feature.
Visualizing a genomic region of interest in GenomeView and exporting the genomic features from a genomic region
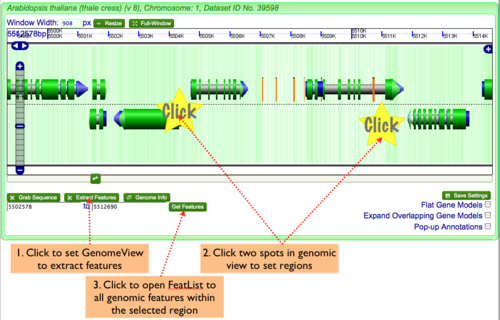
Visualizing a genomic region
After linking to GenomeView from either OrganismView or FeatView, GenomeView will display the genomic region in CoGe's dynamic and interactive genome browser. This tool lets you navigate to the left and right, zoom in and out, and change the types of genomic information displayed. To display the annotation for a particular genomic feature, just click on the feature in the displayed genomic region's graphic. Please see GenomeView for more information.
- Adjust the genomic view by zooming in and out on the genomic region, and scolling the view left and right until the entire genomic region of interest is displayed in the GenomeView.
- Determine the exact boundaries of the region by clicking of genomic features to get their annotations.
Exporting the features of a genomic region in GenomeView to FeatList
To genomic features from GenomeView:
- Click "Extract Features" button in lower left of navigation screen.
- Click two regions in the genomic view to set the start and stop positions of the sequence you wish to grab.
- Click "Get Features" to launch FeatList and get a list of all genomic features in the selected region.
Evaluating, selecting, and exporting genomic features in FeatList
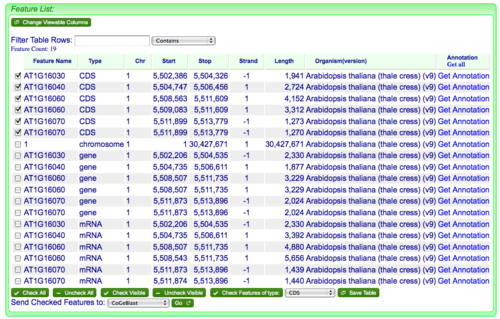
When FeatList loads after being launched from GenomeView, all the genomic features that overlap the selected genomic region are displayed. This usually includes genes, mRNAs, CDSs, and a chromosome, but may include other types of genomic features as well. To select genomic features for export, you can check them or if you want to select and entire type of genomic features, there is a button at the bottom of the list called "Check Features of type:" followed by a drop-down menu of all the listed feature types. Just select the type you want and click the button.
Exporting selected genomic features from FeatList to FastaView
To export the features as fasta sequences, just:
- Open the drop-down menu located next to "Send Checked Features to:" at the bottom of the list and select "FASTA sequences".
- Press the button labeled "Go" next to the drop-down menu.
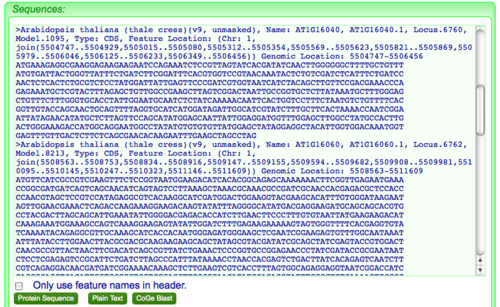
Displaying Fasta formatted sequences in FastaView
Converting nucleotide sequences to protein sequences for CDS features in FastaView
FastaView will generate fasta formatted sequences for a list of genomic features. When it first loads, the CDS sequences will be for DNA, and spliced together appropriately for each exon.
- To convert the sequences to amino acid protein sequences, just click the "Protein Sequence" button displayed below the sequences.
Copying sequences from FastaView and pasting into another application (e.g. text file)
- To export the sequences:
- Double click anywhere in the text area displaying the sequences.
- Or, alternatively, press the the "Plain Text" button.
- Then copy the sequences using by pressing ctrl-c (windows, linux), apple-c (mac), or selecting your web browser's menu Edit->Copy.
- Then paste the sequences into another application, web tool, or text file by pressing ctrl-v (windows, linus), apple-v (mac) or selecting your web browser's menu Edit->Paste.