Computationally Comparing Different Genomes
Background Information
Objective: To have you select a gene in one organism and find where that gene has moved to in another organism. Also have you notice how much the gene has changed.
Difficulty: Easy
Estimated Time: 10 minutes
New Programs Used: FastaView, CoGeBlast, & GEvo
Lesson
Introduction
A human genome contains over 3 billion base pairs. Now, if a scientist wants to compare portions of human DNA to a chimpanzee's DNA, which is also over 3 billion base pairs long, the scientist would need to use a computer program or the task would take hundreds of years. This is where CoGe comes in. CoGe can be used to quickly take a segment of a 3 billion base pair genome and find where that segment closely matches another genome. This process uses an algorithm to do the search. BLAST (Basic Local Alignment Search Tool) is a very commonly used algorithm in genetics for these types of searches. As you finish this lesson you will begin to understand how comparing genetic elements may be used to determine ancestry.
Part 1 - Finding the DNA sequence of the gene you want to compare
1. Open OrganismView (quicklink) and search for the organism you want to have compared and select it. For now, search "Escherichia coli strain" and select the first result
- Note: Escherichia coli is the unabbreviated name for E. coli, a very well studied bacteria.
2. Click Launch Genome Viewer in the bottom left corner to open a new window that visually depicts the genome
3. Click the gene you want to have compared, for now select a random gene (Note: you'll probably want to zoom in to do this).
- A small window titled features should pop up containing information on this gene
4. Click on the link to the right of Location: in the Features window to open a new window that contains the DNA sequence of the gene (quicklink)
5. Click CoGe Blast to open a new web page and to begin the blasting process (quicklink)
Part 2 - Blasting the gene to another genome
You should now be looking at a web page titled CoGeBlast. The large box containing many A's, T's, C's, G's and N's shows the DNA sequence for the gene you previously selected (Note: the N's signify unsequenced DNA). Now you need to find the entire genome of the other organism you want to compare this gene with. To do this:
1. Search for the organism you want to compare your gene with in the Organism Name search box and select it, for now search again for "Escherichia coli strain"
2. Now you are going to compare your orginal gene with all of E. coli strains in the results screen. Click the + Add all listed button under the search results screen
- Note: you just added the original organism you chose to the list of genomes that will be blasted. To remove it, select it in the Genomes to BLAST box then click - Remove
3. Click Run CoGe Blast to compare the gene in the first organism you selected to the entire genome of all the other organisms you selected (quicklink)
4. You just completed the blasting process
Part 3 - Analyzing the BLAST results
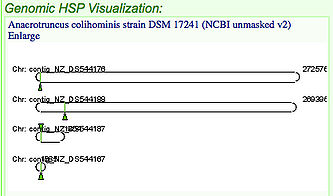
The results page contains a lot of information and one of the first things you'll notice is a picture like the one to the right. This picture shows the location of the similar genes that were found through the blast search by marking them with a green arrow. In the picture to the right four similar genes were found on four different chromosomes and/or organisms. The following steps will show you how to compare the locations of these genes to the location of the original gene you chose.
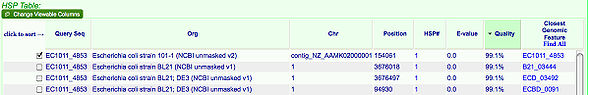
1. Select the top box and any other box in the HSP Table (shown on the right) which has a quality near 97%
Notes:
- The top box is the original gene you selected and the second box you chose is the gene you are comparing it to
- Each row is a gene that was found to be similar by the BLAST and the quality tells us how closely the genes match.
2. Click Go, located right underneath the table. This will open a new window which displays information on the genes you just selected, ignore this information for now and click Run GEvo Analysis!
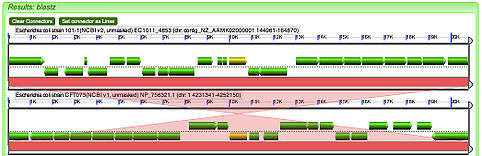
3. The new window that opened displays your final result, two genomes one on top of the other with red bars to mark regions of these genomes that are similar to one another based on their DNA sequence. The genes you selected are displayed as yellow arrows as opposed to the usual green.
Though this result may not seem amazing, the information you just acquired is extremely useful for biologists. Biologists now know where the gene is located in the second organism, the yellow arrow, and how much the genomes have changed, as noted by the quality.
Conclusion
What you just did
Part 1
1. select an organism
2. select a gene in this organism
Part 2
3. choose other organisms to which to compare that gene
Part 3
4. use an algorithm called BLAST to find the most similar genes in organisms you just selected to the gene you selected
5. select the original gene and a blasted gene
6. visually compare the two genes
Biology you should learn from this
Homolog: In biology a Homolog can be described as shared traits between organisms that is related through sharing a common ancestor. For example a human's arm is homologous to a bear's arm because they have a shared ancestry. In genetics, homologs are used to describe two or more genes related through a shared ancestry. For example a gene that codes for Protein A in humans is homologous to the gene for Protein A in bears because the gene was present in an ancient ancestor. Additionally, the gene for Protein A in humans and bears is homologous to the gene for Protein A in their common ancestor.
Ortholog: In genetics, an Ortholog is a special case of homology where genes in two different organisms have originated from a common ancestor. It is often assumed that orthologs have conserved the same function as the ancestoral gene, and hence have similar functions to one another.
Paralog: In genetics a Paralog is a special case of homology when a gene is duplicated within an organism. For example the gene for Protein A is copied so now there are two genes for Protein A in the genome. These genes are paralogs of one another.
- Note: when a gene is copied, one of the genes may mutate (evolve) freely. For the example, one of the two genes may mutate to code for Protein A'. In this case the genes for Protein A and A' are still paralogs, but may have different functions.
- Note: paralogous genes are homologs, but not orthologs. Likewise, orthologous genes are homologs, but not paralogs.
See Also
Previous Lecture: Visually Comparing Genomes
Next Lecture:Synteny: Getting the Big Picture
All Lectures: Tutorial for High School Students