Gene assembly
David Nelson contributed this workflow. His general problem is to find and annotate P450 genes in a new genome (OrgFoo)
Here is a description of my process that could be useful to others.
Contents
Searching a genome using CoGeBlast
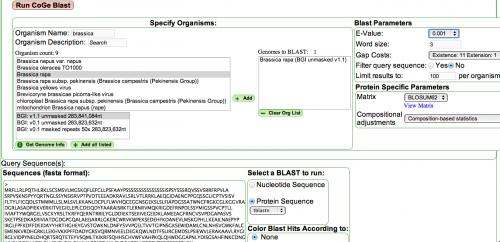
- Use CoGe Blast with a small section of the protein sequence you are working on.
- Set the parameters to search OrgFoo
- add that genome to the "genomes to blast" dialog box with the + add button.
- Set "filter" to off
- set the expect at a high value like 10, 100 for short pieces.
- Note from Eric: CoGeBlast has ways to limit the number of hits. This is set to 100 per organism. You can increase this value but know that having a limit is useful because:
- Many people are interested in the best hits
- Cuts down on the amount of information set to a user's web browser (e.g. Firefox). More data == more the browser has to work.
- Note from Eric: CoGeBlast has ways to limit the number of hits. This is set to 100 per organism. You can increase this value but know that having a limit is useful because:
- word length to 3, (2 for short pieces).
- paste your seq in the FASTA sequences window.
- This requires an identifier line
- I use ">kk" or some thing like that, at least one character is needed since it does not work with > by itself.
- Note by Eric: Thanks for identifying this problem. It has been fixed. ">" as a header will now work.
- select protein to get the tblastn program.
- Click the Run CoG Blast button.
Analyzing Blast Output
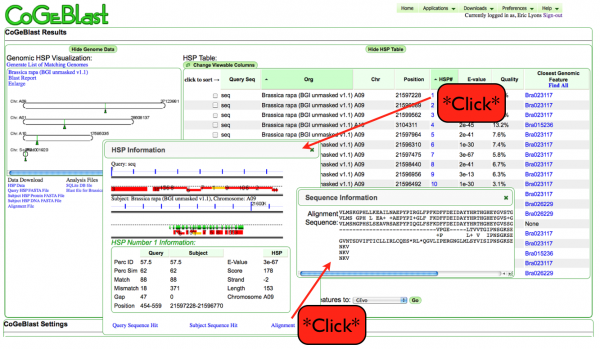
- At the top of the output page, select the appropriate hit (may take some trials to find the right one)
- If you know the scaffold number just try the first entry for that scaffold.
- Click on the HSP number (hyperlinked) to retrieve the blast hit.
- The HSP information box will appear. To see if you have the right sequence click on alignment (lower right)
- This will bring up a box with the blast alignment in it. (it might be under the HSP information box).
- drag the HSP information box out of the way if you have to. Drag the alignment box under the HSP information box so it will not be covered up later. You can leave it there and click on other HSP#s to get new info in the HSP information box and clicking alignment again will give you the new alignmment
- (it is not automatic. You have to click alignment each time).
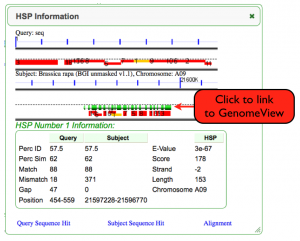
- You can use the HSP information box to get the start and stop location of your sequence.
- The numbers are always presented in the order (left end of sequence, right end of sequence)
- This is true even if the strand is minus, so then the number on the left will be larger than the number on the right.
- This is a good feature, since you do not have to remember the strandedness.
Extracting Sequence with GenomeView
- Once you have found the right sequence to work on, go to the HSP information box and click on the second blue line with tic marks and a kb nucleotide numbering number (NOTE: this is the second genomic panel in the HSP info box).
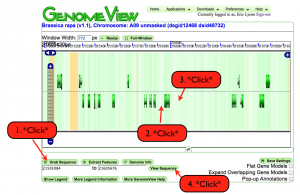
- This will open an other window (GenomeView) with a browser page showing the sequence you blasted in the very center and lots of gene models all around it.
- The green bars are models (usually quite narrow in the default view).
- For more information on these graphics see: GenomeView examples
- here you can get the DNA sequence for the region around your blast hit.
- Click on the grab sequence button (lower left) than move your mouse to the immediate left of the blast hit (in the center of the page) and click once. The nucleotide location enters the window under the grab sequence button.
- Move the mouse to the immediate right of your hit and click once. The location goes into the second window under the garb sequence button.
- Now click the view sequence button. This will give you a new page (SeqView) with the selected sequence range included.
- You can copy and past this into other programs.
Analyze Extracted Sequence
- Here is what I do with it now.
- I have a Do-it-yourself Blast server set up with 474 representative plant P450 sequences in FASTA format pasted into the database window.
- I actually have two of these pages, one for the OrgFoo P450s I am annotating since these are often the best matches to other OrgFoo P450s.
- Paste the Nucleotide sequence from the CoGe site into the query window and do a blastx search against your personal database of sequences.
- The results will come back in few seconds and you can assemble your gene roughly from these blast results.
- To refine the intron exon boundaries paste the CoGe OrgFoo Nucleotide sequence into the Expasy DNA translator
For a first look I usually use the compact option. If I need to see nucleotide sequence to find intron-exon boundaries, then use the "includes nucleotide sequence" option. even if you paste in 50,000 bp of sequence you can jump to the region of interest by using the find command and typing in your amino acid sequence. If the nucleotide translation is on be sure to put two spaces between each amino acid letter. Be aware that the find command cannot wrap at line ends so if you don't find your sequence, search with some sequence to the left or right a few amino acids and this can take you to the right place.
These tools will permit you to assemble any gene.
Suggestions for CoGe
- Small item for improving CoGe.
- Even though I turned the filter off, and the expect value is at 100, I still cannot always get N-terminal low complexity sequence to show up in the blast output. It behaves as if the filter is on. Eric, you might want to check that.
- Additional improvement: The translate option in your sequence window translates in three frames, but they are not interleaved. This makes detection of frameshifts impossible. It would be very very nice to offer a three frame translation with all three frames included (like the JGI three frame nucleotide translation view). If you did that it would be worth a medal.
David